
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,193,285 – 22,193,375 |
| Length | 90 |
| Max. P | 0.710605 |

| Location | 22,193,285 – 22,193,375 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 74.48 |
| Shannon entropy | 0.50211 |
| G+C content | 0.36257 |
| Mean single sequence MFE | -17.20 |
| Consensus MFE | -8.68 |
| Energy contribution | -9.41 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22193285 90 - 27905053 UUGUUGUCGACG--UUUUUUAUUAAUAAAAAUGUGUUGAUAAUUUAAAAGUGCUUUGCACGCGCAUCAUGUGCAUCGAUAAGCCUCUUCCUU------ ...(((((((..--....(((((((((......))))))))).......((((...))))((((.....)))).)))))))...........------ ( -19.80, z-score = -1.82, R) >droSim1.chr3R 22040072 90 - 27517382 UUGUUGUCAACG--UUUUUUAUUAAUAAAAUUGUGUUGAUAAUUUAAAAGUGCUUUGCACGCGCAUCAUGUGCAUCGAUAAGCCCCUUCCUU------ ..((((((((((--..(((((....)))))...))))))))))......((((...))))((((.....))))...................------ ( -18.60, z-score = -1.99, R) >droSec1.super_13 981010 90 - 2104621 UUAUUGUCAACG--UUUUUUAUUAAUAAAAUUGUGUUGAUAAUUUAAAAGUGCUUUGCACGCUCAUCAUGUGCAUCGAUAAGCCCCUUCCUU------ ..((((((((((--..(((((....)))))...))))))))))........((((((((((.......)))))).....)))).........------ ( -16.50, z-score = -2.12, R) >droYak2.chr3R 4544139 92 + 28832112 UUGUUGUCGGCGCUUUUUUUAUUAAUAAAAUUGUGUUGAUAAUUUAAAAGUGCUUUGCACGCGCAUCAUGUGCAUCGAUAAGCCUCUUCCUC------ ...((((((((((((((.(((((((((......)))))))))...))))))))..((((((.......)))))).))))))...........------ ( -25.50, z-score = -3.34, R) >droEre2.scaffold_4820 4620761 89 + 10470090 UUGUUGUCGGCG---UUUUUAUUAAUAAAAUUGUGUUGAUAAUUUAAAAGUGCUUUGCACGCGCAUCAUGUGCAUCGAUAAGCCUCUUUCUC------ ...(((((((..---...(((((((((......))))))))).......((((...))))((((.....)))).)))))))...........------ ( -18.90, z-score = -0.98, R) >droWil1.scaffold_181130 7048168 85 - 16660200 UUUUUCUC-CCG--UUCAUUAUUAAU----UUCUGUUGAUAGUUUAAAAGUGCUUUGCAUGCGCAUCAUGUGCAUCGAUAAGCUUCAUCAUC------ ........-...--...(((((((((----....)))))))))....((((...(((.((((((.....)))))))))...)))).......------ ( -15.90, z-score = -1.61, R) >droVir3.scaffold_13047 4726020 79 - 19223366 -----------GUGUUGCCCU--------UUCCUGUUGAUUAUUUAAAAGUACUUUGCAUUCGCUUCAUGUGCAUCGAUAAGCGCCAUCAUUUCAACG -----------..........--------.....(((((..........((((...((....)).....))))...(((.......)))...))))). ( -9.90, z-score = 0.55, R) >droMoj3.scaffold_6540 12474522 87 + 34148556 -----------GUGCUGUCCUUUUAUUAAUUCCUGCUGAUAAUUUAAAAGUGCUUUGCAAUCGAUUCAUGUACGUCGAUAAGCCCCGUCGUUUCACUG -----------.(((.(..((((((.....((.....)).....))))))..)...)))((((((........))))))................... ( -12.50, z-score = -0.53, R) >consensus UUGUUGUC_ACG__UUUUUUAUUAAUAAAAUUGUGUUGAUAAUUUAAAAGUGCUUUGCACGCGCAUCAUGUGCAUCGAUAAGCCCCUUCCUU______ .................................(((((((.........((((...))))((((.....))))))))))).................. ( -8.68 = -9.41 + 0.74)
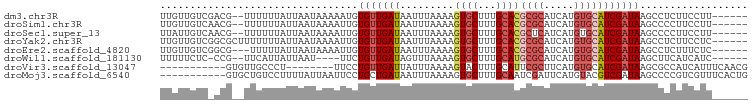
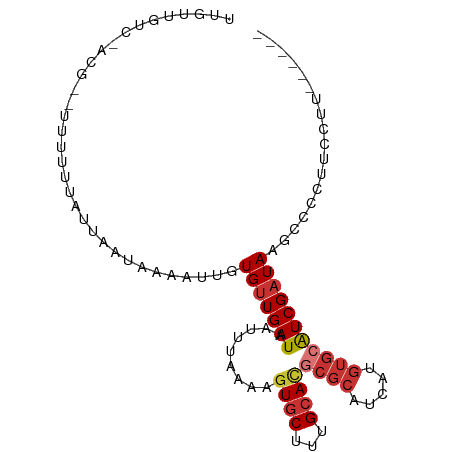

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:24 2011