| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,190,212 – 22,190,302 |
| Length | 90 |
| Max. P | 0.657982 |

| Location | 22,190,212 – 22,190,302 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.43 |
| Shannon entropy | 0.46388 |
| G+C content | 0.39924 |
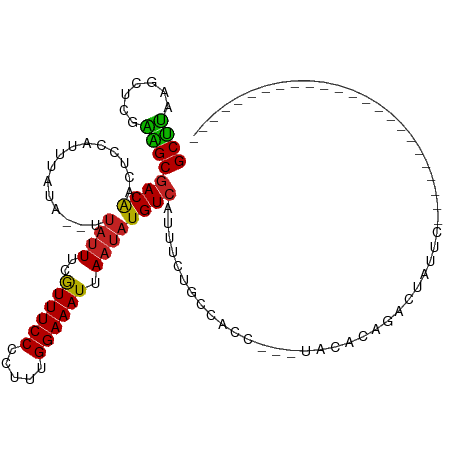
| Mean single sequence MFE | -15.32 |
| Consensus MFE | -9.21 |
| Energy contribution | -8.71 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.657982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
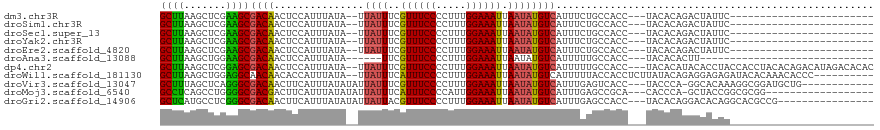
>dm3.chr3R 22190212 90 - 27905053 GCUUAAGCUCGAAGCGACAACUCCAUUUAUA--UUAUUUCGUUUCCCCUUUGGAAAUUAAUAUGUCAUUUCUGCCACC---UACACAGACUAUUC------------------------ ((((.......))))((((..........((--(((....((((((.....)))))))))))))))...((((.....---....))))......------------------------ ( -11.20, z-score = -1.04, R) >droSim1.chr3R 22036975 90 - 27517382 GCUUAAGCUCGAAGCGACAACUCCAUUUAUA--UUAUUUCGUUUCCCCUUUGGAAAUUAAUAUGUCAUUUCUGCCACC---UACACAGACUAUUC------------------------ ((((.......))))((((..........((--(((....((((((.....)))))))))))))))...((((.....---....))))......------------------------ ( -11.20, z-score = -1.04, R) >droSec1.super_13 977970 90 - 2104621 GCUUAAGCUCGAAGCGACAACUCCAUUUAUA--UUAUUUCGUUUCCCCUUUGGAAAUUAAUAUGUCAUUUCUGCCACC---UACACAGACUAUUC------------------------ ((((.......))))((((..........((--(((....((((((.....)))))))))))))))...((((.....---....))))......------------------------ ( -11.20, z-score = -1.04, R) >droYak2.chr3R 4540999 90 + 28832112 GCUUAAGCUCGAAGCGACAACUCCAUUUAUA--UUAUUUCGUUUCCCCUUUGGAAAUUAAUAUGUCAUUUCUGCCACC---UACACAGACUAUUC------------------------ ((((.......))))((((..........((--(((....((((((.....)))))))))))))))...((((.....---....))))......------------------------ ( -11.20, z-score = -1.04, R) >droEre2.scaffold_4820 4617747 90 + 10470090 GCUUAAGCUCGAAGCGACAACUCCAUUUAUA--UUAUUUCGUUUCCCCUUUGGAAAUUAAUAUGUCAUUUCUGCCACC---UACACAGACUAUUC------------------------ ((((.......))))((((..........((--(((....((((((.....)))))))))))))))...((((.....---....))))......------------------------ ( -11.20, z-score = -1.04, R) >droAna3.scaffold_13088 72754 81 - 569066 GCUUAAGCUGGAAGCGACAACUCCAUUUAUA------UUCGUUUCCCCUUUGGAAAUUAAUAUGUCAUUUUUGCCACC---UACACACUU----------------------------- ((....)).((..((((..........((((------((.((((((.....)))))).))))))......))))..))---.........----------------------------- ( -12.09, z-score = -1.26, R) >dp4.chr2 11989802 114 + 30794189 GCUUAAGCUCGGAGCGACAACUCCAUUUAUA--UUAUUUCGUUUCCCCUUUGGAAAUUAAUAUGUCAUUUUUGCCACC---UACACAUACACCUACCACCUACACAGACAUAGACACAC ((....))..((((......)))).......--.......((((((.....))))))...((((((............---.........................))))))....... ( -13.49, z-score = -1.17, R) >droWil1.scaffold_181130 9677585 107 + 16660200 GCUUAAGCUGGAGGCAACAACACCAUUUAUA--UUAUUUCAUUUCCCCUUUGGAAAUUAAUAUGUCAUUUUUACCACCUCUUAUACAGAGGAGAGAUACACAAACACCC---------- ((....)).((.(....).............--.((((((((((((.....))))))...................(((((.....))))).))))))........)).---------- ( -16.90, z-score = -1.36, R) >droVir3.scaffold_13047 4722989 103 - 19223366 GCUUUAGCUCAGGGCGACAACUUCAUUUAUAUAUUAUUUCGUUUCCCCUUUGGAAAUUAAUAUGUCAUUUGAGUCACC---UACCCA-GGCACAAAGGCGGAUGCUG------------ (((((.(((..(((.(((....(((..(((((((((....((((((.....)))))))))))))).)..)))))).))---).....-)))...)))))........------------ ( -21.00, z-score = -0.36, R) >droMoj3.scaffold_6540 12470835 97 + 34148556 GCCUCAGCCUGGGGCGACGACUUCAUUUAUAUAUUAUUUCAUUUCCCCAUUGGAAAUUAAUAUGUCAUUUGAGCCGCA---CACCCA-GCUACCGGCGCGG------------------ (((..(((.(((((.(.((.((.((..(((((((((....((((((.....)))))))))))))).)..)))).))).---).))))-)))...)))....------------------ ( -26.40, z-score = -1.99, R) >droGri2.scaffold_14906 10163873 100 - 14172833 GCUCAUGCCUCGGGCGACAACUUCAUUUAUAUAUUAUUACGUUUCCCCUUUGGAAAUUAAUAUGUCAUUUGAGCCACC---UACACAGGACACAGGCACGCCG---------------- ((....))....((((......(((..(((((((((....((((((.....)))))))))))))).)..)))(((.((---(....))).....))).)))).---------------- ( -22.60, z-score = -2.07, R) >consensus GCUUAAGCUCGAAGCGACAACUCCAUUUAUA__UUAUUUCGUUUCCCCUUUGGAAAUUAAUAUGUCAUUUCUGCCACC___UACACAGACUAUUC________________________ ((((.......))))((((...............((((..((((((.....)))))).))))))))..................................................... ( -9.21 = -8.71 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:23 2011