
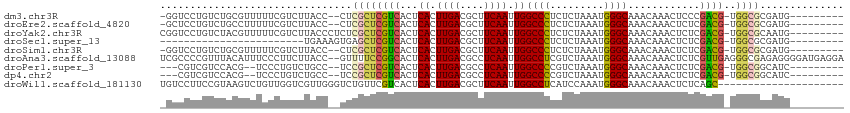
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,163,738 – 22,163,839 |
| Length | 101 |
| Max. P | 0.973555 |

| Location | 22,163,738 – 22,163,839 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.05 |
| Shannon entropy | 0.56735 |
| G+C content | 0.54032 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -14.63 |
| Energy contribution | -14.72 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
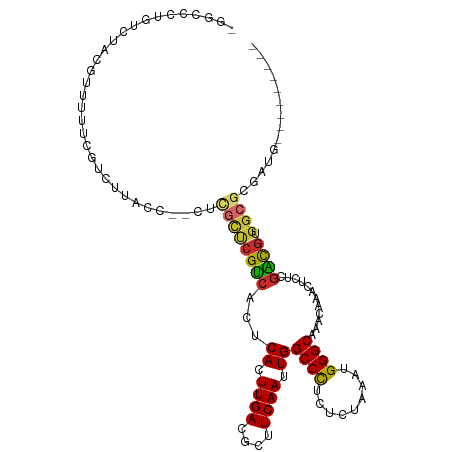
>dm3.chr3R 22163738 101 - 27905053 -GGUCCUGUCUGCGUUUUUCGUCUUACC--CUCGCUCGUCACUCACUUGACGCUUCAAUUGGCCCUCUCUAAAUGGGCAAACAAACUCCCGACG-UGGCGCGAUG--------- -......(((.(((((...((((.....--......(((((......))))).........((((.........))))............))))-.)))))))).--------- ( -25.20, z-score = -1.42, R) >droEre2.scaffold_4820 4587880 101 + 10470090 -GCUCCUGUCUGCCUUUUUCGUCUUACC--CUCGCUCGUCACUCACUUGACGCUUCAAUUGGCCCUCUCUAAAUGGGCAAACAAACUCUCGACG-UGGCGCGAUG--------- -......(((((((.....((((.....--......(((((......))))).........((((.........))))............))))-.)))).))).--------- ( -23.60, z-score = -2.15, R) >droYak2.chr3R 4513677 104 + 28832112 CGGUCCUGUCUACGUUUUUCGUCUUACCCUCUCGCUCGUCACUCACUUGACGCUUCAAUUGGCCCUCUCUAAAUGGGCAAACAAACUCUCGACG-UGGCGCAAUG--------- ......((((((((((....................(((((......))))).........((((.........))))............))))-))).)))...--------- ( -20.90, z-score = -0.89, R) >droSec1.super_13 951285 80 - 2104621 ------------------------UGAAAGUGAGCUCGUCACUCACUUGACGCUUCAAUUGGCCCUCUCUAAAUGGGCAAACAAACUCUCGACG-UGGCGCGAUG--------- ------------------------.....(((..(.((((...((.((((....)))).))((((.........))))............))))-.).)))....--------- ( -18.80, z-score = -0.54, R) >droSim1.chr3R 22010136 101 - 27517382 -GGUCCUGUCUGCGUUUUUCGUCUUACC--CUCGCUCGUCACUCACUUGACGCUUCAAUUGGCCCUCUCUAAAUGGGCAAACAAACUCUCGACG-UGGCGCGAUG--------- -......(((.(((((...((((.....--......(((((......))))).........((((.........))))............))))-.)))))))).--------- ( -25.20, z-score = -1.60, R) >droAna3.scaffold_13088 43555 112 - 569066 UCGCCCCGUUUACAUUUCCCUUCUUACC--GUUUUCCGGCACUCACUUGACGCCUCAAUUGGCCUCGUCUAAAUGGGCAAACAAACUCUCGUUGAGGGCGAGAGGGGAUGAGGA .....(((....)...((((((((..((--((((((((.........(((.(((......))).)))......)))).))))...(((.....)))))..))))))))...)). ( -32.46, z-score = -0.13, R) >droPer1.super_3 5814093 97 + 7375914 ---CGUCGUCCACG--UCCCUGUCUGCC--UCCGCUCGUCACUCACUUGACGCCUCAAUUGGCCCCGUCUAAAUGGGCAAACAAACUCUCGACG-UGGCGGCAUC--------- ---.((((.(((((--((..(((.((((--(......((((......))))(((......)))...........))))).))).......))))-)))))))...--------- ( -33.60, z-score = -3.89, R) >dp4.chr2 11956725 97 + 30794189 ---CGUCGUCCACG--UCCCUGUCUGCC--UCCGCUCGUCACUCACUUGACGCCUCAAUUGGCCCCGUCUAAAUGGGCAAACAAACUCUCGACG-UGGCGGCAUC--------- ---.((((.(((((--((..(((.((((--(......((((......))))(((......)))...........))))).))).......))))-)))))))...--------- ( -33.60, z-score = -3.89, R) >droWil1.scaffold_181130 9650107 93 + 16660200 UGUCCUUCCGUAAGUCUGUUGGUCGUUGGGUCUGUUCGUCACUCACUUGACGCUUCAAUUGGCCUCAUCCAAAUGGGCAAACAAACUCUCAGC--------------------- (((((...(....)....((((....((((((.((((((((......)))))....))).)))).)).))))..)))))..............--------------------- ( -19.90, z-score = -0.21, R) >consensus _GGCCCUGUCUACGUUUUUCGUCUUACC__CUCGCUCGUCACUCACUUGACGCUUCAAUUGGCCCUCUCUAAAUGGGCAAACAAACUCUCGACG_UGGCGCGAUG_________ ................................((((((((...((.((((....)))).))((((.........))))............))))..)))).............. (-14.63 = -14.72 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:19 2011