
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,155,710 – 22,155,806 |
| Length | 96 |
| Max. P | 0.584683 |

| Location | 22,155,710 – 22,155,806 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 76.76 |
| Shannon entropy | 0.45009 |
| G+C content | 0.40457 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -14.51 |
| Energy contribution | -14.18 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22155710 96 + 27905053 -----------UUUGGCUGCAA-GCGAGAG-------CUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCAGUC-----AGAGCUGGCCAAAAAAG-CCA -----------(((((((((((-((....)-------))....((((((.((((...((((.........))))...)))).)))))))))))))-----)))...(((.......)-)). ( -27.40, z-score = -2.05, R) >droGri2.scaffold_14906 10126932 87 + 14172833 ----------UUUUGCCAGCAUUGCAAGCG-----AGCUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCUGCUGGCCCCG------------------- ----------....((((((...((.....-----.))..(((((((((.((((...((((.........))))...)))).))))).))))))))))....------------------- ( -22.80, z-score = -1.64, R) >droMoj3.scaffold_6540 12433450 100 - 34148556 ----------UUUUGCCAGCACUGCAAGCG-----AGCUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCUGCCGGCCCGGGGUACGGCUGCGG------ ----------...(((.(((..(((...((-----.((((.((((.............((((....)))).(((((((.....))))))))))))))).))..)))..)))))).------ ( -26.80, z-score = -0.00, R) >droVir3.scaffold_13047 4680405 100 + 19223366 ----------UUUUGCUAGCACUGCAAGCG-----AGCUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCUGCCAGCCCAGGGUCCAAGUACGG------ ----------..(((((.((...)).))))-----)((((.((((.............((((....)))).(((((((.....))))))))))))))).................------ ( -22.00, z-score = 0.07, R) >droAna3.scaffold_13088 34418 98 + 569066 -----------UUUGGCUGCAA-GCGAA---------CUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCAGCCCGGGCAGAGC-AGCCGAAAAAGCCA- -----------(((((((((..-.....---------....((((((((.((((...((((.........))))...)))).))))).))).((....))...))-))))))).......- ( -27.20, z-score = -1.85, R) >droEre2.scaffold_4820 4579819 98 - 10470090 -----------UUUGGCUGCAA-GCGAGCG-----AGCUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCAGUC-----AGAGCUGGCCAAAAAAG-CCA -----------..(((((....-((.(((.-----..((((((((((((.((((...((((.........))))...)))).))))).)))..))-----)).))).))......))-))) ( -27.40, z-score = -1.58, R) >droYak2.chr3R 4505314 98 - 28832112 -----------UUUGGCUGCAA-GCGAGAG-----AGCUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCAGUC-----AGAGCUGGCCAAAAAAG-CCA -----------(((((((((((-((.....-----.)))....((((((.((((...((((.........))))...)))).)))))))))))))-----)))...(((.......)-)). ( -25.60, z-score = -1.24, R) >droSec1.super_13 943329 97 + 2104621 -----------UUUGGCUGCAA-GCGAGAG-------CUGAGCAAAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCAGUC-----AGAGCUGGCCAAAAAAAACCA -----------(((((((((.(-((....)-------))..)))...........................(((((((.....)))))))((((.-----...))))))))))........ ( -23.40, z-score = -1.38, R) >droSim1.chr3R 22001954 96 + 27517382 -----------UUUGGCUGCAA-GCGAGAG-------CUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCAGUC-----AGAGCUGGCCAAAAAAG-CCA -----------(((((((((((-((....)-------))....((((((.((((...((((.........))))...)))).)))))))))))))-----)))...(((.......)-)). ( -27.40, z-score = -2.05, R) >droPer1.super_3 5805384 93 - 7375914 -----------GAUGGCUGCAA-GCGAG---------CUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCAGCC-----GGAGC-GGCCAAAAAAGGCA- -----------..(((((((..-.((.(---------(((...((((((.((((...((((.........))))...)))).))))))..)))))-----)..))-))))).........- ( -27.10, z-score = -1.89, R) >dp4.chr2 11948053 93 - 30794189 -----------GAUGGCUGCAA-GCGAG---------CUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCAGCC-----GGAGC-GGCCAAAAAAGGCC- -----------..(((((((((-((...---------....)).(((((.((((...((((.........))))...)))).)))))))))))))-----)....-((((......))))- ( -27.90, z-score = -2.25, R) >droWil1.scaffold_181130 9641495 106 - 16660200 GUUGCCUUAUUUUCGUUGGUAUCGCAAGAGCUGCGAGCUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCAGGCCA---AAAACGCAA------------ ...((((........(..((.(((((.....)))))))..)((((((((.((((...((((.........))))...)))).))))).)))))))..---.........------------ ( -22.50, z-score = 0.01, R) >consensus ___________UUUGGCUGCAA_GCGAGAG_______CUGAGCAGAAAUUAAUUACAUUGAAGUCAUUUAUCAAGAAAGUUUAUUUCUUGCAGCC_____AGAGCUGGCCAAAAAAG_CC_ ..............(((((((..((.((.........))..))((((((.((((...((((.........))))...)))).))))))))))))).......................... (-14.51 = -14.18 + -0.34)

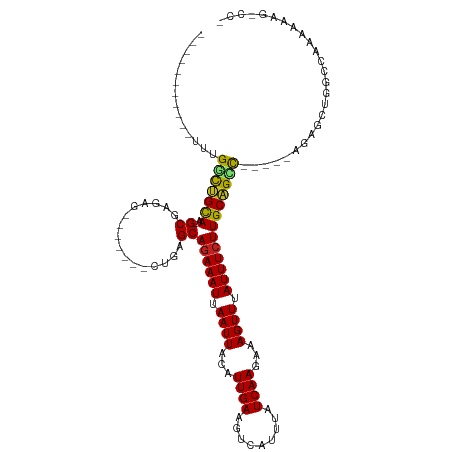
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:18 2011