
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,138,619 – 22,138,709 |
| Length | 90 |
| Max. P | 0.985084 |

| Location | 22,138,619 – 22,138,709 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 66.69 |
| Shannon entropy | 0.65189 |
| G+C content | 0.42583 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -10.89 |
| Energy contribution | -10.98 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
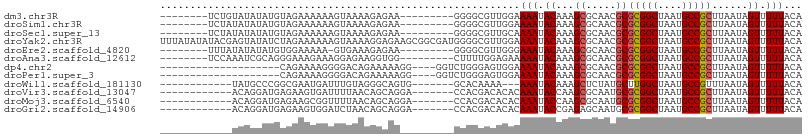
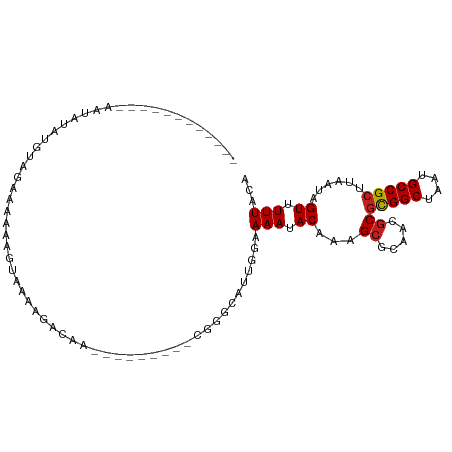
>dm3.chr3R 22138619 90 + 27905053 --------UCUGUAUAUAUGUAGAAAAAAGUAAAAGAGAA---------GGGGCGUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA --------((((........)))).....(((((((..((---------(.((((((((..........((((...))))..)))))))).))).....))))))). ( -24.60, z-score = -2.35, R) >droSim1.chr3R 21983231 90 + 27517382 --------UCUAUAUAUAUGUAGAAAAAAGUAAAAGAGAA---------GGGGCGUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA --------((((((....)))))).....(((((((..((---------(.((((((((..........((((...))))..)))))))).))).....))))))). ( -27.20, z-score = -3.68, R) >droSec1.super_13 926093 90 + 2104621 --------UCUAUAUAUAUGUAGAAAAAAGUAAAAGAGAA---------GGGGCGUUGCAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA --------((((((....)))))).....(((((((....---------...(((((((............)))))))(((((....))))).......))))))). ( -29.10, z-score = -4.14, R) >droYak2.chr3R 4487686 107 - 28832112 UUUAUAUAUACGAGUAUAUCUAGAAAAAAGUAAAAGGAGAAGCGGCGAUGGGGCGUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA ((((.(((((....))))).)))).....(((((((...((((.(((.((..(((((...........)))))..)))))(((....))))))).....))))))). ( -29.40, z-score = -2.21, R) >droEre2.scaffold_4820 4562445 89 - 10470090 --------UUUAUAUAUAUGUGGAAAAA-GUGAAAGAGAA---------GGGGCGUUGGGAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA --------((((((....))))))....-(((((((....---------...((((((.(..........).))))))(((((....))))).......))))))). ( -24.50, z-score = -2.38, R) >droAna3.scaffold_12612 28207 90 + 30757 --------UCCAAAUCGCAGGGAAAGAAAGGAGAAGGUGG---------CUUUUGGAGAAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA --------((((((((((..................))))---------..)))))).((((((.....(((...)))(((((....)))))......))))))... ( -19.67, z-score = 0.24, R) >dp4.chr2 11928719 83 - 30794189 --------------------CAGAAAAGGGGACAGAAAAAGG----GGUCUGGGAGUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA --------------------............((((......----..))))...(((((((.......(((...)))(((((....))))).......))))))). ( -20.10, z-score = -1.46, R) >droPer1.super_3 5785049 83 - 7375914 --------------------CAGAAAAGGGGACAGAAAAAGG----GGUCUGGGAGUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA --------------------............((((......----..))))...(((((((.......(((...)))(((((....))))).......))))))). ( -20.10, z-score = -1.46, R) >droWil1.scaffold_181130 9614808 85 - 16660200 ------------UAUGCCCGGCGAAUGAUUUGUAGGGCAGUG-------GCACAAAA---AAAUACAAAGCUCUAUGCUUGGCUAAUGCCGUUUAAUAGUUUUUACA ------------..(((((.((((.....)))).)))))...-------......((---((((...((((.....))))(((....)))........))))))... ( -21.00, z-score = -0.45, R) >droVir3.scaffold_13047 14562279 88 - 19223366 ------------ACAGGAUGAGAAGUGAUUUUAACAGCAGGA-------CCACGACACACAAAUACCAAGCGCAAUGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA ------------............((((..(((..(((....-------....................((((...))))(((....))))))...)))...)))). ( -15.70, z-score = 0.04, R) >droMoj3.scaffold_6540 21733524 88 + 34148556 ------------ACAGGAUGAGAAGCGGUUUUAACAGCAGGA-------CCACGACACACAAAUACCAAGCGCAAUGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA ------------..(((((...((((((((((......))))-------))..................((((...))))(((....)))))))....))))).... ( -20.50, z-score = -1.15, R) >droGri2.scaffold_14906 4063704 88 - 14172833 ------------ACAGGAUGAGAAGUGGAUCUAACAGCAGGA-------CCACGACACACAAAUACCGAGAGCAAUGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA ------------......((.(..((((.(((.......)))-------))))..).))........((((((.((..(((((....)))))...)).))))))... ( -21.10, z-score = -1.87, R) >consensus ____________AAUAUAUGUAGAAAAAAGUAAAAGACAA_________CGGGCAUUGGAAAAUACAAAGCGCAACGCGCGGCUAAUGCCGCUUAAUAGUUUUUACA ............................................................(((.((...((.....))(((((....)))))......)).)))... (-10.89 = -10.98 + 0.09)
| Location | 22,138,619 – 22,138,709 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 66.69 |
| Shannon entropy | 0.65189 |
| G+C content | 0.42583 |
| Mean single sequence MFE | -18.37 |
| Consensus MFE | -10.08 |
| Energy contribution | -10.33 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
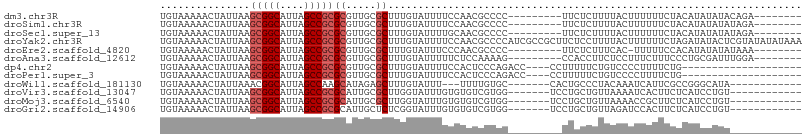
>dm3.chr3R 22138619 90 - 27905053 UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUCCAACGCCCC---------UUCUCUUUUACUUUUUUCUACAUAUAUACAGA-------- .((((((........(((((....)))))((((((..............))))))...---------.....)))))).....................-------- ( -19.44, z-score = -2.54, R) >droSim1.chr3R 21983231 90 - 27517382 UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUCCAACGCCCC---------UUCUCUUUUACUUUUUUCUACAUAUAUAUAGA-------- .((((((........(((((....)))))((((((..............))))))...---------.....)))))).....................-------- ( -19.44, z-score = -2.51, R) >droSec1.super_13 926093 90 - 2104621 UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUGCAACGCCCC---------UUCUCUUUUACUUUUUUCUACAUAUAUAUAGA-------- .((((((........(((((....)))))((((((((..........))))))))...---------.....)))))).....................-------- ( -24.10, z-score = -3.57, R) >droYak2.chr3R 4487686 107 + 28832112 UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUCCAACGCCCCAUCGCCGCUUCUCCUUUUACUUUUUUCUAGAUAUACUCGUAUAUAUAAA .((((((........(((((....)))))((((((..............)))))).................))))))..........(((((....)))))..... ( -20.74, z-score = -1.35, R) >droEre2.scaffold_4820 4562445 89 + 10470090 UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUCCCAACGCCCC---------UUCUCUUUCAC-UUUUUCCACAUAUAUAUAAA-------- (((............(((((....)))))((((((..............))))))...---------...........-.......)))..........-------- ( -16.54, z-score = -2.12, R) >droAna3.scaffold_12612 28207 90 - 30757 UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUUCUCCAAAAG---------CCACCUUCUCCUUUCUUUCCCUGCGAUUUGGA-------- ...............(((((....)))))(((...)))............((((((.(---------(.....................))..))))))-------- ( -16.20, z-score = 0.10, R) >dp4.chr2 11928719 83 + 30794189 UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUCCACUCCCAGACC----CCUUUUUCUGUCCCCUUUUCUG-------------------- ...............(((((....)))))(((...))).................((((..----......))))............-------------------- ( -14.00, z-score = -1.49, R) >droPer1.super_3 5785049 83 + 7375914 UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUCCACUCCCAGACC----CCUUUUUCUGUCCCCUUUUCUG-------------------- ...............(((((....)))))(((...))).................((((..----......))))............-------------------- ( -14.00, z-score = -1.49, R) >droWil1.scaffold_181130 9614808 85 + 16660200 UGUAAAAACUAUUAAACGGCAUUAGCCAAGCAUAGAGCUUUGUAUUU---UUUUGUGC-------CACUGCCCUACAAAUCAUUCGCCGGGCAUA------------ .................(((....)))..((((((((..........---))))))))-------...(((((...............)))))..------------ ( -16.46, z-score = 0.12, R) >droVir3.scaffold_13047 14562279 88 + 19223366 UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCAUUGCGCUUGGUAUUUGUGUGUCGUGG-------UCCUGCUGUUAAAAUCACUUCUCAUCCUGU------------ ..............(((((((...((((((...(((((.........))))).)))))-------)..)))))))....................------------ ( -20.00, z-score = -0.41, R) >droMoj3.scaffold_6540 21733524 88 - 34148556 UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCAUUGCGCUUGGUAUUUGUGUGUCGUGG-------UCCUGCUGUUAAAACCGCUUCUCAUCCUGU------------ ..............(((((((...((((((...(((((.........))))).)))))-------)..)))))))....................------------ ( -20.00, z-score = 0.13, R) >droGri2.scaffold_14906 4063704 88 + 14172833 UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCAUUGCUCUCGGUAUUUGUGUGUCGUGG-------UCCUGCUGUUAGAUCCACUUCUCAUCCUGU------------ ........(((....(((((....)))))(((..(((..((..((....))..)).))-------)..)))...)))..................------------ ( -19.50, z-score = -0.42, R) >consensus UGUAAAAACUAUUAAGCGGCAUUAGCCGCGCGUUGCGCUUUGUAUUUUCCAACCCCCC_________UUCCCUUUUACUUUUUUCUACAUAUAUA____________ ...............(((((....)))))((.....))..................................................................... (-10.08 = -10.33 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:16 2011