| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,119,586 – 22,119,680 |
| Length | 94 |
| Max. P | 0.613508 |

| Location | 22,119,586 – 22,119,680 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.37 |
| Shannon entropy | 0.48147 |
| G+C content | 0.56880 |
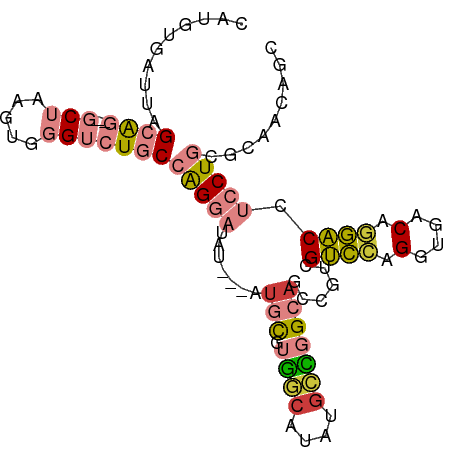
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.21 |
| Covariance contribution | -0.17 |
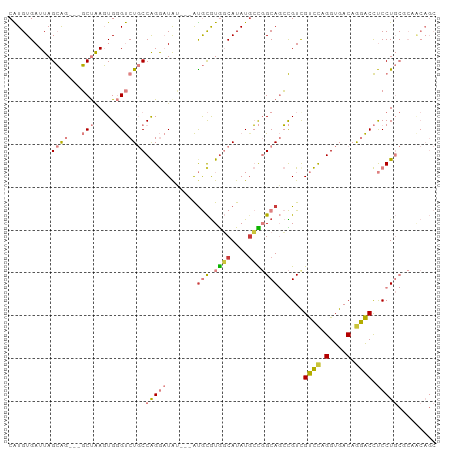
| Combinations/Pair | 1.44 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
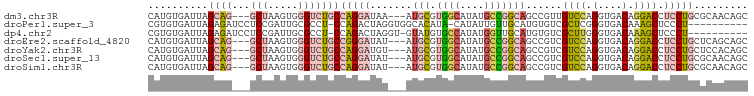
>dm3.chr3R 22119586 94 - 27905053 CAUGUGAUUAGCAG---GCUAAGUGGGUCUGCCAGGAUAA---AUGCGUGGCAUAUGCCGGCAGCCGUUGUCCAGGUGACAGGACCUCCUGCGCAACAGC ..(((.....((((---(......(((((((((.((((((---.(((.((((....)))))))....)))))).)))....)))))))))))...))).. ( -35.90, z-score = -1.07, R) >droPer1.super_3 5765533 88 + 7375914 CGUGUGAUUAGAGAUCCUCCGAUUGCGCCU-CCAGACUAGGUGGCACAUA-CAUAUUGUUGCAUGUGUCGCUCGGGUGACAAAGCUCCCU---------- .(((..((..(((...)))..))..)))..-........((((((((((.-((......)).)))))))))).(((.(......).))).---------- ( -28.10, z-score = -1.28, R) >dp4.chr2 11907995 88 + 30794189 CGUGUGAUUAGAGAUCCUCCGAUUGCGCCU-CCAGACUAGGU-GUAUGUGCCAUAUGGUUGCAUGUGUCGCUUGGGUGACAAAGCUCCCU---------- .(((..((..(((...)))..))..)))..-........((.-.((((((((....)).))))))..))((((.(....).)))).....---------- ( -26.40, z-score = -0.89, R) >droEre2.scaffold_4820 4541036 94 + 10470090 CAUGUGAUUAGCAG---GCUAAGUGGGUCUGCCGGGAUAU---AUGCGUGGCAUAUGCCGGCAGCCGUCGUCCAGGUGACAGGACCUCCUGCUCAGCAGC ..(((....(((((---(.......((.(((((((..(((---((((...))))))))))))))))...((((.(....).))))..))))))..))).. ( -40.70, z-score = -1.88, R) >droYak2.chr3R 4468345 94 + 28832112 CAUGUGAUUAGCAG---GCUAAGUGGGUCUGCCAGGAUGU---AUGCGUGGCAUAUGCCGGCAGCCGUCGUCCAGGUGACAGGACCUCCUGCUCCACAGC ..((((...(((((---(.......((.(((((.((.(((---((((...))))))))))))))))...((((.(....).))))..)))))).)))).. ( -39.60, z-score = -1.62, R) >droSec1.super_13 906928 94 - 2104621 CAUGUGAUUAGCAG---GCUAAGUGGGUCUGCCAGGAUAU---AUGCGUGGCAUAUGCCGGCAGCCGUCGUCCAGGUGACAGGACCUCCUGCGCAACAGC ..(((.....((((---(.......((.(((((.((.(((---((((...))))))))))))))))...((((.(....).))))..)))))...))).. ( -38.00, z-score = -1.50, R) >droSim1.chr3R 21963612 94 - 27517382 CAUGUGAUUAGCAG---GCUAAGUGGGUCUGCCAGGAUAU---AUGCGUGGCAUAUGCCGGCAGCCGUCGUCCAGGUGACAGGACCUCCUGCGCAACAGC ..(((.....((((---(.......((.(((((.((.(((---((((...))))))))))))))))...((((.(....).))))..)))))...))).. ( -38.00, z-score = -1.50, R) >consensus CAUGUGAUUAGCAG___GCUAAGUGGGUCUGCCAGGAUAU___AUGCGUGGCAUAUGCCGGCAGCCGUCGUCCAGGUGACAGGACCUCCUGCGCAACAGC .......(((((.....)))))..(((((((((.((((......(((.((((....)))))))......)))).)))....))))))............. (-18.39 = -18.21 + -0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:15 2011