
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,114,175 – 22,114,280 |
| Length | 105 |
| Max. P | 0.657941 |

| Location | 22,114,175 – 22,114,280 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.32 |
| Shannon entropy | 0.25788 |
| G+C content | 0.43208 |
| Mean single sequence MFE | -32.01 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.06 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.657941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22114175 105 - 27905053 -------------CUGACCAUG-UGCAAUCGCCUGAUAUUUGGUGGUUGCCUCGGUCGC-UUUAUGACUGCACUGGGACCAAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAGCAAAGUG -------------..((((..(-.(((((((((........))))))))))..))))((-(((..(((.....((....))...(((((((((....)))))))))))))))))...... ( -34.60, z-score = -3.39, R) >droSim1.chr3R 21958191 105 - 27517382 -------------CUGACCAUG-UGCAAUCGCCUGAUAUUUGGUGGUUGCCUCGGUCGC-UUUAUGACUGCACUGGGACCAAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAGCAAAGUG -------------..((((..(-.(((((((((........))))))))))..))))((-(((..(((.....((....))...(((((((((....)))))))))))))))))...... ( -34.60, z-score = -3.39, R) >droSec1.super_13 894166 105 - 2104621 -------------CUGACCAUG-UGCAAUCGCCUGAUAUUUGGUGGUUGCCUCGGUCGC-UUUAUGACUGCACUGGGACCAAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAGCAAAGUG -------------..((((..(-.(((((((((........))))))))))..))))((-(((..(((.....((....))...(((((((((....)))))))))))))))))...... ( -34.60, z-score = -3.39, R) >droYak2.chr3R 4462824 105 + 28832112 -------------CUGACCAUG-UGCAAUCGCCUGAUAUUUGGUGGUUGCCUCGGUCGC-UUUAUGACUGCACUGGGACCAAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAGCAAAGUG -------------..((((..(-.(((((((((........))))))))))..))))((-(((..(((.....((....))...(((((((((....)))))))))))))))))...... ( -34.60, z-score = -3.39, R) >droEre2.scaffold_4820 4535743 105 + 10470090 -------------CUGACCAUG-UGCAAUCGCCUGAUAUUUGGUGGUUGCCUCGGUCGC-UUUAUGACUGCACUGGGACCAAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAGCAAAGUG -------------..((((..(-.(((((((((........))))))))))..))))((-(((..(((.....((....))...(((((((((....)))))))))))))))))...... ( -34.60, z-score = -3.39, R) >droAna3.scaffold_13340 23655102 105 - 23697760 -------------CUGGCCAUA-UGCAAUCGUCUGAUAUUUGGUGGUCGGGUCGGUCGC-UUUAUGACUGCACUGGGACCAAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAGCAAAGUG -------------((((((((.-....(((....))).....))))))))..((((((.-....))))))((((..(.......(((((((((....))))))))).......)..)))) ( -27.04, z-score = -0.58, R) >dp4.chr2 11902895 106 + 30794189 -------------CUGACCAUA-UGCAAUCGCCUGAUAUUUGGUGGUUGCCUCGGUCACUUUUAUGACUGCACUGGGACCCAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAGCAAAGUG -------------.(((((...-.(((((((((........)))))))))...)))))............((((..(.......(((((((((....))))))))).......)..)))) ( -31.04, z-score = -2.46, R) >droPer1.super_3 5760450 106 + 7375914 -------------CUGACCAUA-UGCAAUCGCCUGAUAUUUGGUGGUUGCCUCGGUCACUUUUAUGACUGCACUGGGACCCAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAGUAAAGUG -------------.(((((...-.(((((((((........)))))))))...)))))........(((..(((..(((.....(((((((((....))))))))))))..)))..))). ( -32.70, z-score = -3.25, R) >droWil1.scaffold_181130 6958887 97 - 16660200 -------------UGGCCCAUA-UGCAAUCGUGCGAUAUUUGGUGGCU-UCUUGGUCAC-UUUAUGACUGCAUGGGGAGCCAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAG------- -------------((((((.((-((((.(((((........(((((((-....))))))-).))))).)))))).)).))))..(((((((((....))))))))).......------- ( -32.40, z-score = -2.85, R) >droVir3.scaffold_13047 4626716 105 - 19223366 -------------CUGGCCAUA-UGCAAUCGUGCGAUAUUUGGUGGCUUCCUCGGUCAC-UUUAUGACUGCACUGGGACCAAUAAUUUAUGAAUGAGUUCAUGAAUGUCGAAGCUGAUUC -------------...((((..-((((....)))).....))))((((((..(((((.(-(...((....))..))))))....(((((((((....))))))))))..))))))..... ( -26.80, z-score = -0.61, R) >droMoj3.scaffold_6540 12381532 105 + 34148556 -------------CUGGCUAUA-UGCAAUCGUGUGAUAUUUGGUGGCUGCGUCGGUCAC-UUUAUGACUGCACUGGGAGCAAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAGCUGAUUC -------------..((((...-(((..(((((((.(((..((((((((...)))))))-)..))).).)))).))..)))...(((((((((....))))))))).....))))..... ( -27.80, z-score = -1.18, R) >droGri2.scaffold_14906 10067129 119 - 14172833 UCAGCCUUUUGUGUCGCUUGCAGUGCAAUCGUGCGAUAUUUGCUGGUUGCUCCGGUCAC-UUUAUGACUGCGCUGGGACCAAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAACUGAUUC ((((....(((.(((.((.((.(.(((((((.((((...))))))))))).)((((((.-....)))))).)).))))))))..(((((((((....))))))))).......))))... ( -33.30, z-score = -1.45, R) >consensus _____________CUGACCAUA_UGCAAUCGCCUGAUAUUUGGUGGUUGCCUCGGUCAC_UUUAUGACUGCACUGGGACCAAUAAUUUAUGAAUGAAUUCAUGAAUGUCGAAGCAAAGUG ...............((((((......(((....))).....)))))).((.((((((......))))))....))(((.....(((((((((....))))))))))))........... (-17.47 = -17.06 + -0.41)

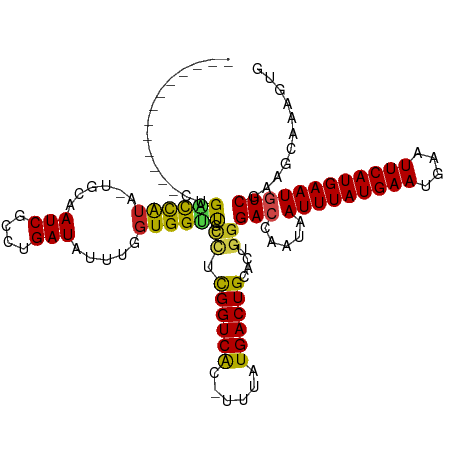
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:13 2011