
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,093,631 – 22,093,721 |
| Length | 90 |
| Max. P | 0.825923 |

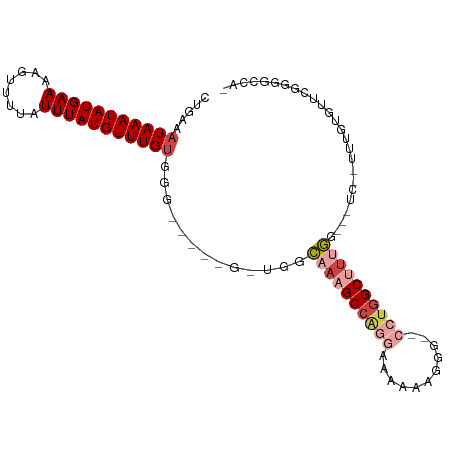
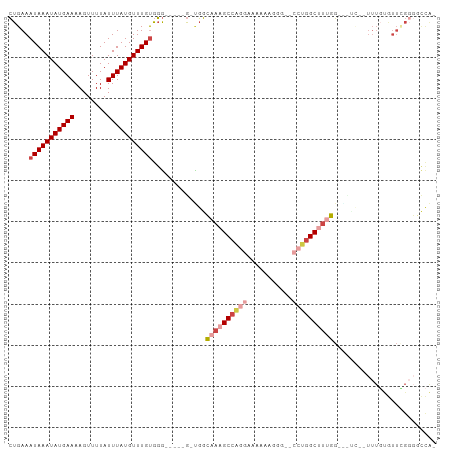
| Location | 22,093,631 – 22,093,721 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 70.70 |
| Shannon entropy | 0.55952 |
| G+C content | 0.43779 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -13.29 |
| Energy contribution | -14.90 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.825923 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
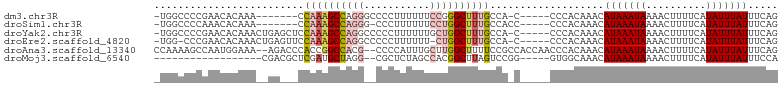
>dm3.chr3R 22093631 90 + 27905053 CUGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGUGGG-----G-UGGCAAAGCCCGGAAAAAAGGGGCCCUGGCUUUGG-------UUUGUGUUCGGGGCCA- .....(((((((((((........))))))))))).((-----(-(......))))..........(((((((((.....-------.....).)))))))).- ( -26.50, z-score = -1.19, R) >droSim1.chr3R 21937756 90 + 27517382 CUGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGUGGG-----GGUGGCAAAGCCAGGAAAAAAGGG-CCCUGGCUUUGG-------UUUGUGUUUGGGGCCA- (..(((((((((((((........)))))))))))...-----.....((((((((((.........-.)))))))))).-------......))..).....- ( -24.40, z-score = -0.91, R) >droYak2.chr3R 4441864 97 - 28832112 CUGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGUGGG-----G-UGGCAAAGCCAGCAAAAAAGGGGGCCUGGCUUUGGAGCUCAGUUUGUGUUCGGGGCCA- ((((((((((((((((........))))))))))).((-----(-(..((((((((((..........).)))))))))..))))........))))).....- ( -28.60, z-score = -1.31, R) >droEre2.scaffold_4820 4515053 95 - 10470090 CUGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGUGGG-----G-UGGCAAAGCCAG-AAAAAAGGGGGCCUGGCUUUGGAACUCAGUUUGUGUUCGGG-CCA- ((((((((((((((((........))))))))))).((-----(-(..(((((((((-............)))))))))..))))........))))).-...- ( -28.60, z-score = -2.20, R) >droAna3.scaffold_13340 23628885 100 + 23697760 CUGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGUGGGUUGGUGGCGGAAAAGCCAAGCAAAUGGGG--CGUGGCCGGUGGGUCU--UUUCCAUUGGCUUUUGG (..(.(((((((((((........)))))))))))...)..)((((......)))).((.......)--)..((((((((((...--..))))))))))..... ( -29.00, z-score = -1.90, R) >droMoj3.scaffold_6540 12356421 79 - 34148556 UGGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGCCAC-----CCGGACUAAGCCGUGGCUAGAGCG--CCUAGCAUCGAGCGUCG------------------ (((...((((((((((........))))))))))))).-----...(((...((((..(((((....--.)))))..)).))))).------------------ ( -18.20, z-score = -0.44, R) >consensus CUGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGUGGG_____G_UGGCAAAGCCAGGAAAAAAGGG__CCUGGCUUUGG___UC__UUUGUGUUCGGGGCCA_ .....(((((((((((........))))))))))).............((((((((((...........))))))))))......................... (-13.29 = -14.90 + 1.61)

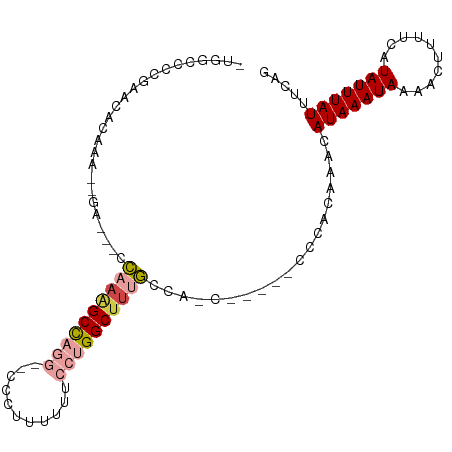
| Location | 22,093,631 – 22,093,721 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 70.70 |
| Shannon entropy | 0.55952 |
| G+C content | 0.43779 |
| Mean single sequence MFE | -16.01 |
| Consensus MFE | -7.93 |
| Energy contribution | -9.07 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745372 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 22093631 90 - 27905053 -UGGCCCCGAACACAAA-------CCAAAGCCAGGGCCCCUUUUUUCCGGGCUUUGCCA-C-----CCCACAAACAUAAAUAAAACUUUUCAUAUUUAUUUCAG -.......(((......-------.(((((((.(((.........))).)))))))...-.-----.........(((((((..........)))))))))).. ( -12.80, z-score = 0.22, R) >droSim1.chr3R 21937756 90 - 27517382 -UGGCCCCAAACACAAA-------CCAAAGCCAGGG-CCCUUUUUUCCUGGCUUUGCCACC-----CCCACAAACAUAAAUAAAACUUUUCAUAUUUAUUUCAG -((((............-------..((((((((((-........))))))))))))))..-----.........(((((((..........)))))))..... ( -17.53, z-score = -2.50, R) >droYak2.chr3R 4441864 97 + 28832112 -UGGCCCCGAACACAAACUGAGCUCCAAAGCCAGGCCCCCUUUUUUGCUGGCUUUGCCA-C-----CCCACAAACAUAAAUAAAACUUUUCAUAUUUAUUUCAG -................(((((...(((((((((.............)))))))))...-.-----.........(((((((..........)))))))))))) ( -15.82, z-score = -0.89, R) >droEre2.scaffold_4820 4515053 95 + 10470090 -UGG-CCCGAACACAAACUGAGUUCCAAAGCCAGGCCCCCUUUUUU-CUGGCUUUGCCA-C-----CCCACAAACAUAAAUAAAACUUUUCAUAUUUAUUUCAG -(((-(..((((.(.....).)))).(((((((((..........)-))))))))))))-.-----.........(((((((..........)))))))..... ( -18.20, z-score = -2.47, R) >droAna3.scaffold_13340 23628885 100 - 23697760 CCAAAAGCCAAUGGAAA--AGACCCACCGGCCACG--CCCCAUUUGCUUGGCUUUUCCGCCACCAACCCACAAACAUAAAUAAAACUUUUCAUAUUUAUUUCAG ....((((.(((((...--.(......)(((...)--))))))).))))(((......)))..............(((((((..........)))))))..... ( -14.80, z-score = -1.44, R) >droMoj3.scaffold_6540 12356421 79 + 34148556 ------------------CGACGCUCGAUGCUAGG--CGCUCUAGCCACGGCUUAGUCCGG-----GUGGCAAACAUAAAUAAAACUUUUCAUAUUUAUUUCCA ------------------...((((((..((((((--(((....))....))))))).)))-----)))......(((((((..........)))))))..... ( -16.90, z-score = -0.36, R) >consensus _UGGCCCCGAACACAAA__GA___CCAAAGCCAGG__CCCUUUUUUCCUGGCUUUGCCA_C_____CCCACAAACAUAAAUAAAACUUUUCAUAUUUAUUUCAG .........................((((((((((...........))))))))))...................(((((((..........)))))))..... ( -7.93 = -9.07 + 1.14)
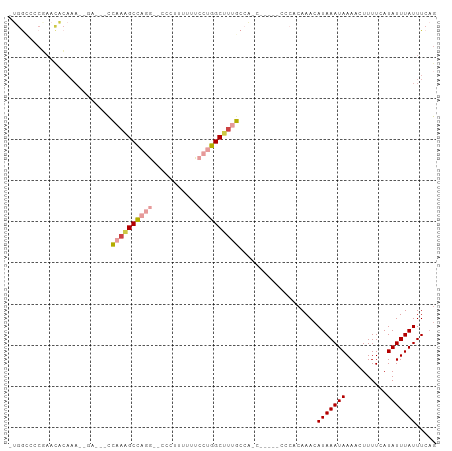
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:12 2011