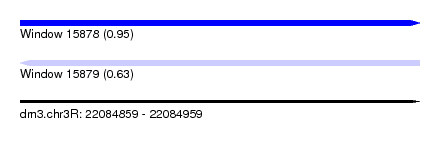
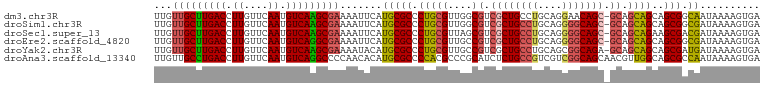
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,084,859 – 22,084,959 |
| Length | 100 |
| Max. P | 0.953487 |

| Location | 22,084,859 – 22,084,959 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 87.38 |
| Shannon entropy | 0.23963 |
| G+C content | 0.55069 |
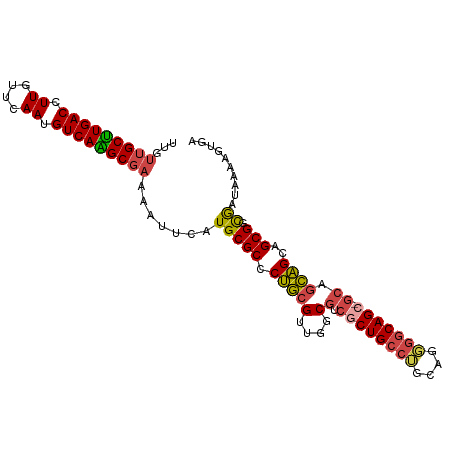
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -29.19 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
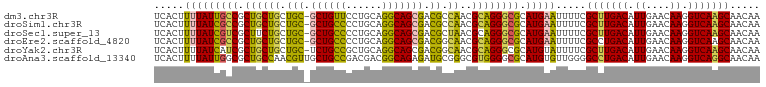
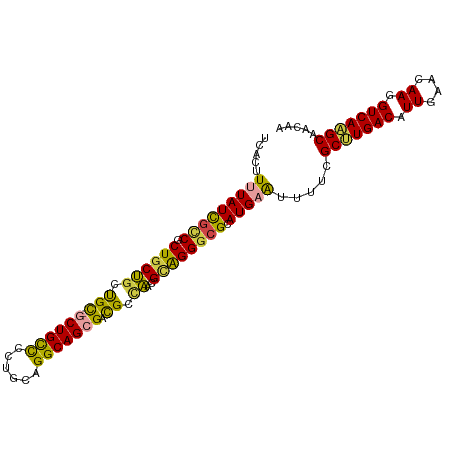
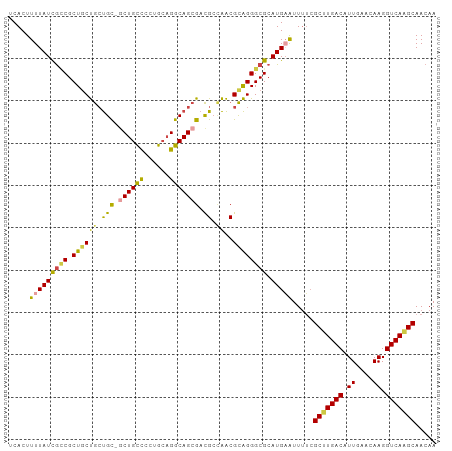
>dm3.chr3R 22084859 100 + 27905053 UCACUUUUAUUGCCGCUGCUGCUGC-GCUGUUCCUGCAGGCAGCGACGCCAACGCAGGGCGCAUGAAUUUUCGCUUGACAUUGAACAAGGUCAAGCAACAA .....(((((((((.((((.(((((-.((((....)))))))))(....)...)))))))).))))).....(((((((.((....)).)))))))..... ( -36.70, z-score = -1.64, R) >droSim1.chr3R 21929367 100 + 27517382 UCACUUUUAUCGCCGCUGCUGCUGC-GCUGCCCCUGCAGGCAGCGACGCCAACGCAGGGCGCAUGAAUUUUCGCUUGACAUUGAACAAGGUCAAGCAACAA .....(((((((((.((((.((..(-((((((......)))))))..))....)))))))).))))).....(((((((.((....)).)))))))..... ( -41.80, z-score = -3.15, R) >droSec1.super_13 864773 100 + 2104621 UCACUUUUAUCGUCGCUUCUGCUGC-GCUGCCCCUGCAGGCAGCGACGCUAACGCAGGGCGCAUGAAUUUUCGCUUGACAUUGAACAAGGUCAAGCAACAA .........(((((((((.((((((-.((((....))))))))))..((....)).))))).))))......(((((((.((....)).)))))))..... ( -36.90, z-score = -2.43, R) >droEre2.scaffold_4820 4506054 100 - 10470090 UCACUUUUAUCGCCGCUGCUGCUGC-GCUGCCCCUGCAGGCAGCGACGGCAACGCAGGGCGCAUGAAUUUUCGCCUGACAUUGAACAAGGUCAAGCAACAA .....(((((((((.((((((((((-((((((......))))))).)))))..)))))))).))))).....((.((((.((....)).)))).))..... ( -43.40, z-score = -3.24, R) >droYak2.chr3R 4432765 100 - 28832112 UCACUUUUAUCAUCGCUGCUGCUGC-UCUGCCGCUGCAGGCAGCGACGGCAACGCAGGGCGCAUGUAUUUUCGCUUGACAUUGAACAAGGUCAAGCAACAA ................((((((.((-((((((((((....)))))..(....))))))))))).))).....(((((((.((....)).)))))))..... ( -37.20, z-score = -2.10, R) >droAna3.scaffold_13340 23620591 101 + 23697760 UCACUUUUAUUGGCGCUGCCAACGUUGCUGCCGACGACGGCAGAGAUGCGGGCGUGGGGCGCAUGUGUUGGGGCCUGACAUUGAACAAGGUCAGGCAACAA ............((((((((..(((..((((((....))))))..)))..)))....))))).....(((..(((((((.((....)).)))))))..))) ( -43.40, z-score = -2.58, R) >consensus UCACUUUUAUCGCCGCUGCUGCUGC_GCUGCCCCUGCAGGCAGCGACGCCAACGCAGGGCGCAUGAAUUUUCGCUUGACAUUGAACAAGGUCAAGCAACAA .....(((((((((.((((((((((..((((....))))))))))..(....))))))))).))))).....(((((((.((....)).)))))))..... (-29.19 = -29.62 + 0.42)
| Location | 22,084,859 – 22,084,959 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 87.38 |
| Shannon entropy | 0.23963 |
| G+C content | 0.55069 |
| Mean single sequence MFE | -39.09 |
| Consensus MFE | -29.63 |
| Energy contribution | -29.50 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22084859 100 - 27905053 UUGUUGCUUGACCUUGUUCAAUGUCAAGCGAAAAUUCAUGCGCCCUGCGUUGGCGUCGCUGCCUGCAGGAACAGC-GCAGCAGCAGCGGCAAUAAAAGUGA ...(((((((((.((....)).)))))))))....((((..(((.......)))((((((((((((.........-))))..)))))))).......)))) ( -36.10, z-score = -0.63, R) >droSim1.chr3R 21929367 100 - 27517382 UUGUUGCUUGACCUUGUUCAAUGUCAAGCGAAAAUUCAUGCGCCCUGCGUUGGCGUCGCUGCCUGCAGGGGCAGC-GCAGCAGCAGCGGCGAUAAAAGUGA ...(((((((((.((....)).)))))))))....((((.((((((((....((..((((((((....)))))))-)..)).)))).))))......)))) ( -44.90, z-score = -2.33, R) >droSec1.super_13 864773 100 - 2104621 UUGUUGCUUGACCUUGUUCAAUGUCAAGCGAAAAUUCAUGCGCCCUGCGUUAGCGUCGCUGCCUGCAGGGGCAGC-GCAGCAGAAGCGACGAUAAAAGUGA ...(((((((((.((....)).)))))))))....((((........((((.((.(((((((((((....)))).-))))).)).))))))......)))) ( -35.64, z-score = -0.85, R) >droEre2.scaffold_4820 4506054 100 + 10470090 UUGUUGCUUGACCUUGUUCAAUGUCAGGCGAAAAUUCAUGCGCCCUGCGUUGCCGUCGCUGCCUGCAGGGGCAGC-GCAGCAGCAGCGGCGAUAAAAGUGA ...(((((((((.((....)).)))))))))....((((.((((((((..(((.(.((((((((....)))))))-)).))))))).))))......)))) ( -44.90, z-score = -2.23, R) >droYak2.chr3R 4432765 100 + 28832112 UUGUUGCUUGACCUUGUUCAAUGUCAAGCGAAAAUACAUGCGCCCUGCGUUGCCGUCGCUGCCUGCAGCGGCAGA-GCAGCAGCAGCGAUGAUAAAAGUGA ...(((((((((.((....)).))))))))).....(((.(((..((((((((((((((((....))))))).).-))))).))))))))).......... ( -39.20, z-score = -2.25, R) >droAna3.scaffold_13340 23620591 101 - 23697760 UUGUUGCCUGACCUUGUUCAAUGUCAGGCCCCAACACAUGCGCCCCACGCCCGCAUCUCUGCCGUCGUCGGCAGCAACGUUGGCAGCGCCAAUAAAAGUGA (((..(((((((.((....)).)))))))..)))((((((((.........)))))..((((((....))))))....((((((...))))))....))). ( -33.80, z-score = -1.76, R) >consensus UUGUUGCUUGACCUUGUUCAAUGUCAAGCGAAAAUUCAUGCGCCCUGCGUUGGCGUCGCUGCCUGCAGGGGCAGC_GCAGCAGCAGCGGCGAUAAAAGUGA ...(((((((((.((....)).))))))))).......(((((.(((((....)((.(((((((....))))))).)).))))..))).)).......... (-29.63 = -29.50 + -0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:10 2011