
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,078,790 – 22,078,911 |
| Length | 121 |
| Max. P | 0.824621 |

| Location | 22,078,790 – 22,078,911 |
|---|---|
| Length | 121 |
| Sequences | 11 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Shannon entropy | 0.47364 |
| G+C content | 0.43750 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -16.90 |
| Energy contribution | -17.57 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.604579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
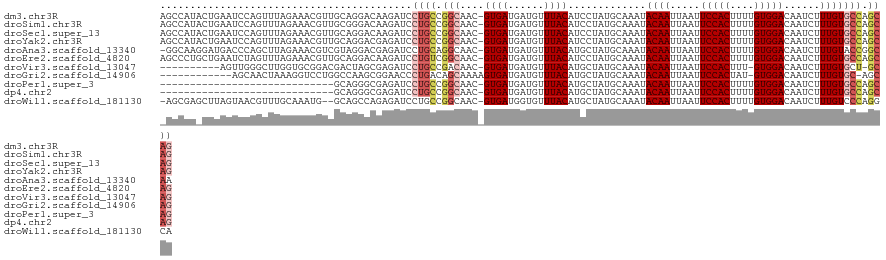
>dm3.chr3R 22078790 121 + 27905053 AGCCAUACUGAAUCCAGUUUAGAAACGUUGCAGGACAAGAUCCUGCCGGCAAC-GUGAUGAUGUUUACAUCCUAUGCAAAUACAAUUAAUUCCACUUUUGUGGACAAUCUUUGUGCCAGCAG .(((..((((....))))...........((((((.....)))))).)))...-.....((((....))))...(((...(((((.....(((((....)))))......)))))...))). ( -31.60, z-score = -1.98, R) >droSim1.chr3R 21922897 121 + 27517382 AGCCAUACUGAAUCCAGUUUAGAAACGUUGCGGGACAAGAUCCUGCCGGCAAC-GUGAUGAUGUUUACAUCCUAUGCAAAUACAAUUAAUUCCACUUUUGUGGACAAUCUUUGUGCCAGCAG ......((((....)))).(((..(((((((.((.((......)))).)))))-))...((((....)))))))(((...(((((.....(((((....)))))......)))))...))). ( -32.70, z-score = -2.01, R) >droSec1.super_13 858728 121 + 2104621 AGCCAUACUGAAUCCAGUUUAGAAACGUUGCAGGACAAGAUCCUGCCGGCAAC-GUGAUGAUGUUUACAUCCUAUGCAAAUACAAUUAAUUCCACUUUUGUGGACAAUCUUUGUGCCAGCAG .(((..((((....))))...........((((((.....)))))).)))...-.....((((....))))...(((...(((((.....(((((....)))))......)))))...))). ( -31.60, z-score = -1.98, R) >droYak2.chr3R 4426507 121 - 28832112 AGCCAUACUGAAUCCAGUUUAGAAACGUUGCAGGACGAGAUCCUGCCGGCAAC-GUGAUGAUGUUUACAUCCUAUGCAAAUACAAUUAAUUCCACUUUUGUGGACAAUCUUUGUGCCAGCAG .(((..((((....))))...........((((((.....)))))).)))...-.....((((....))))...(((...(((((.....(((((....)))))......)))))...))). ( -31.60, z-score = -1.88, R) >droAna3.scaffold_13340 23613745 120 + 23697760 -GGCAAGGAUGACCCAGCUUAGAAACGUCGUAGGACGAGAUCCUGCAGGCAAC-GUGAUGAUGUUUACAUGCUAUGCAAAUACAAUUAAUUCCACUUUUGUGGACAAUCUUUGUACCGGCAA -.((((((((.....(((....(((((((((((((.....)))))).(....)-.....)))))))....))).................(((((....)))))..))))))))........ ( -34.90, z-score = -2.09, R) >droEre2.scaffold_4820 4499920 121 - 10470090 AGCCCUGCUGAAUCUAGUUUAGAAACGUUGCAGGACAAGAUCCUGUCGGCAAC-GUGAUGAUGUUUACAUCCUAUGCAAAUACAAUUAAUUCCACUUUUGUGGACAAUCUUUGUGCCAGCAG ....((((((......((.(((..(((((((..((((......)))).)))))-))...((((....))))))).))...(((((.....(((((....)))))......))))).)))))) ( -38.30, z-score = -3.71, R) >droVir3.scaffold_13047 4574935 109 + 19223366 ----------AGUUGGGCUUGGUGCGGACGACUAGCGAGAUCCUGCCGACAAC-GUGAUGAUGUUUACAUGCUAUGCAAAUACAAUUAAUUCCACUUU-GUGGACAAUCUUUGUGCU-GCAG ----------.(((((((.....))(((((.....))...)))..)))))...-((((......))))......((((..(((((.....(((((...-)))))......))))).)-))). ( -23.40, z-score = 1.15, R) >droGri2.scaffold_14906 10021437 108 + 14172833 ------------AGCAACUAAAGGUCCUGGCCAAGCGGAACCCUGACAGCAAAAGUGAUGAUGUUUACAUGCUAUGCAAAUACAAUUAAUUCCACUAU-GUGGACAAUCUUUGUGC-AGCAG ------------((((......(((.(((......))).)))............((((......)))).)))).(((...(((((.....(((((...-)))))......))))).-.))). ( -20.60, z-score = 1.23, R) >droPer1.super_3 5720086 92 - 7375914 -----------------------------GCAGGGCGAGAUCCUGCCGGCAAC-GUGAUGAUGUUUACAUGCUAUGCAAAUACAAUUAAUUCCACUUUUGUGGACAAUCUUUGUGCCAGCAG -----------------------------((((((.....)))))).((((..-((((......)))).)))).(((...(((((.....(((((....)))))......)))))...))). ( -25.20, z-score = -0.96, R) >dp4.chr2 11862727 92 - 30794189 -----------------------------GCAGGGCGAGAUCCUGCCGGCAAC-GUGAUGAUGUUUACAUGCUAUGCAAAUACAAUUAAUUCCACUUUUGUGGACAAUCUUUGUGCCAGCAG -----------------------------((((((.....)))))).((((..-((((......)))).)))).(((...(((((.....(((((....)))))......)))))...))). ( -25.20, z-score = -0.96, R) >droWil1.scaffold_181130 6911841 118 + 16660200 -AGCGAGCUUAGUAACGUUUGCAAAUG--GCAGCCAGAGAUCCUGCCGGCAAC-GUGAUGGUGUUUACAUGCUAUGCAAAUACAAUUAAUUCCACUUUUGUGGACAAUCUUUGUCCCAGGCA -.....((((.(..(((((((((...(--((((.........)))))((((..-((((......)))).)))).))))))).........(((((....)))))........))..))))). ( -31.30, z-score = -0.22, R) >consensus _GCCAU_CUGAAUCCAGUUUAGAAACGUUGCAGGACGAGAUCCUGCCGGCAAC_GUGAUGAUGUUUACAUGCUAUGCAAAUACAAUUAAUUCCACUUUUGUGGACAAUCUUUGUGCCAGCAG ..........................................((((.(((....((((......)))).............((((.....(((((....)))))......))))))).)))) (-16.90 = -17.57 + 0.67)

| Location | 22,078,790 – 22,078,911 |
|---|---|
| Length | 121 |
| Sequences | 11 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Shannon entropy | 0.47364 |
| G+C content | 0.43750 |
| Mean single sequence MFE | -29.86 |
| Consensus MFE | -22.05 |
| Energy contribution | -22.56 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.824621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
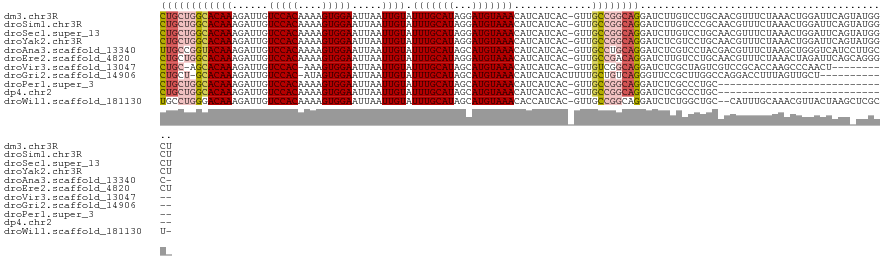
>dm3.chr3R 22078790 121 - 27905053 CUGCUGGCACAAAGAUUGUCCACAAAAGUGGAAUUAAUUGUAUUUGCAUAGGAUGUAAACAUCAUCAC-GUUGCCGGCAGGAUCUUGUCCUGCAACGUUUCUAAACUGGAUUCAGUAUGGCU .((((((((....((((((((((....)))))..))))).....))).(((((((....))))...((-(((((.(((((....))).)).))))))).......)))....)))))..... ( -33.40, z-score = -1.36, R) >droSim1.chr3R 21922897 121 - 27517382 CUGCUGGCACAAAGAUUGUCCACAAAAGUGGAAUUAAUUGUAUUUGCAUAGGAUGUAAACAUCAUCAC-GUUGCCGGCAGGAUCUUGUCCCGCAACGUUUCUAAACUGGAUUCAGUAUGGCU .((((((((....((((((((((....)))))..))))).....))).(((((((....))))...((-(((((.(((((....))).)).))))))).......)))....)))))..... ( -33.40, z-score = -1.33, R) >droSec1.super_13 858728 121 - 2104621 CUGCUGGCACAAAGAUUGUCCACAAAAGUGGAAUUAAUUGUAUUUGCAUAGGAUGUAAACAUCAUCAC-GUUGCCGGCAGGAUCUUGUCCUGCAACGUUUCUAAACUGGAUUCAGUAUGGCU .((((((((....((((((((((....)))))..))))).....))).(((((((....))))...((-(((((.(((((....))).)).))))))).......)))....)))))..... ( -33.40, z-score = -1.36, R) >droYak2.chr3R 4426507 121 + 28832112 CUGCUGGCACAAAGAUUGUCCACAAAAGUGGAAUUAAUUGUAUUUGCAUAGGAUGUAAACAUCAUCAC-GUUGCCGGCAGGAUCUCGUCCUGCAACGUUUCUAAACUGGAUUCAGUAUGGCU ...((((((....((((((((((....)))))..))))).....))).)))((((....)))).....-...(((((((((((...)))))))...........((((....)))).)))). ( -33.20, z-score = -1.38, R) >droAna3.scaffold_13340 23613745 120 - 23697760 UUGCCGGUACAAAGAUUGUCCACAAAAGUGGAAUUAAUUGUAUUUGCAUAGCAUGUAAACAUCAUCAC-GUUGCCUGCAGGAUCUCGUCCUACGACGUUUCUAAGCUGGGUCAUCCUUGCC- .(((..((((((......(((((....))))).....))))))..)))..(((.((((.(........-))))).))).(((...((((....))))..)))..((.(((....))).)).- ( -31.20, z-score = -1.25, R) >droEre2.scaffold_4820 4499920 121 + 10470090 CUGCUGGCACAAAGAUUGUCCACAAAAGUGGAAUUAAUUGUAUUUGCAUAGGAUGUAAACAUCAUCAC-GUUGCCGACAGGAUCUUGUCCUGCAACGUUUCUAAACUAGAUUCAGCAGGGCU (((((((((....((((((((((....)))))..))))).....))).(((((((....))))...((-(((((.(((((....)))))..))))))).......)))....)))))).... ( -38.80, z-score = -3.29, R) >droVir3.scaffold_13047 4574935 109 - 19223366 CUGC-AGCACAAAGAUUGUCCAC-AAAGUGGAAUUAAUUGUAUUUGCAUAGCAUGUAAACAUCAUCAC-GUUGUCGGCAGGAUCUCGCUAGUCGUCCGCACCAAGCCCAACU---------- .(((-((.((((......(((((-...))))).....))))..)))))..((.(((..(((.(.....-).)))..)))((((..(....)..)))))).............---------- ( -21.10, z-score = 0.71, R) >droGri2.scaffold_14906 10021437 108 - 14172833 CUGCU-GCACAAAGAUUGUCCAC-AUAGUGGAAUUAAUUGUAUUUGCAUAGCAUGUAAACAUCAUCACUUUUGCUGUCAGGGUUCCGCUUGGCCAGGACCUUUAGUUGCU------------ .....-((((((((...((((.(-(.(((((((((........(((.((((((.((..........))...))))))))))))))))))))....)))))))).).))).------------ ( -23.10, z-score = 1.11, R) >droPer1.super_3 5720086 92 + 7375914 CUGCUGGCACAAAGAUUGUCCACAAAAGUGGAAUUAAUUGUAUUUGCAUAGCAUGUAAACAUCAUCAC-GUUGCCGGCAGGAUCUCGCCCUGC----------------------------- ((((((((((((......(((((....))))).....)))).(((((((...))))))).........-...)))))))).............----------------------------- ( -27.00, z-score = -1.70, R) >dp4.chr2 11862727 92 + 30794189 CUGCUGGCACAAAGAUUGUCCACAAAAGUGGAAUUAAUUGUAUUUGCAUAGCAUGUAAACAUCAUCAC-GUUGCCGGCAGGAUCUCGCCCUGC----------------------------- ((((((((((((......(((((....))))).....)))).(((((((...))))))).........-...)))))))).............----------------------------- ( -27.00, z-score = -1.70, R) >droWil1.scaffold_181130 6911841 118 - 16660200 UGCCUGGGACAAAGAUUGUCCACAAAAGUGGAAUUAAUUGUAUUUGCAUAGCAUGUAAACACCAUCAC-GUUGCCGGCAGGAUCUCUGGCUGC--CAUUUGCAAACGUUACUAAGCUCGCU- .((.((((((...((((((((((....)))))..)))))...((((((..((((((..........))-).))).(((((.........))))--)...)))))).))).))).)).....- ( -26.90, z-score = 0.81, R) >consensus CUGCUGGCACAAAGAUUGUCCACAAAAGUGGAAUUAAUUGUAUUUGCAUAGCAUGUAAACAUCAUCAC_GUUGCCGGCAGGAUCUCGUCCUGCAACGUUUCUAAACUGGAUUCAG_AUGGC_ ((((((((((((......(((((....))))).....)))).(((((((...))))))).............)))))))).......................................... (-22.05 = -22.56 + 0.51)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:07 2011