
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 22,065,928 – 22,066,113 |
| Length | 185 |
| Max. P | 0.981226 |

| Location | 22,065,928 – 22,066,022 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 79.13 |
| Shannon entropy | 0.37254 |
| G+C content | 0.28540 |
| Mean single sequence MFE | -20.91 |
| Consensus MFE | -12.64 |
| Energy contribution | -14.00 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
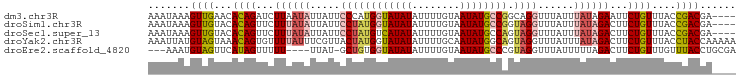
>dm3.chr3R 22065928 94 + 27905053 AAAUAAAGUUGAACACAGAUCUUAAUAUUAUUCCCAUGGUAUAUAUUUUGUAAUAUGCCGGCAGGUUUAUUUAUAGAAUUCUGUUUACCGACGA---- .......((((...(((((..((.(((.((..((..((((((((........))))))))...))..))..))).))..)))))....))))..---- ( -19.30, z-score = -1.79, R) >droSim1.chr3R 21909550 94 + 27517382 AAAUAAAGUUGUACACAGUUCUUUAUAUUAUUCCUAUGGUAUAUAUUUUGUAAUAUGCCGGUAGGUUUAUUUAUAGACUUCUGUUUACCGACGA---- .......((((((.((((...((((((.((..((((((((((((........))))))).)))))..))..))))))...)))).)).))))..---- ( -24.30, z-score = -3.44, R) >droSec1.super_13 845825 94 + 2104621 AAAUAAAGUUGUACACAGUUCUUUAUAUUAUUCCUAUGUCAUAUAUUUUGUAAUAUGCCAGUAGGUUUAUUUAUAGACUUCUGUUUACCGACGA---- .......((((((.((((...((((((.((..(((((..(((((........)))))...)))))..))..))))))...)))).)).))))..---- ( -20.10, z-score = -2.81, R) >droYak2.chr3R 4413487 98 - 28832112 AAAUUAUGUAGUAAACAGUGUUUUAUUUCGUUACUAUGGUAUAUAUUUUGCAAUAUGGCAGUAGGUUUAUUUAUAGACUUCUGUUUACCUACCAAAAA .......(((((((((((.((((..........(((((.(((((........))))).)).)))..........))))..))))))).))))...... ( -18.25, z-score = -0.92, R) >droEre2.scaffold_4820 4483673 90 - 10470090 ---AAAUGUAGUUCAUAGUUUUU----UUAU-GCUGUGGUAUAUAUUUUGUAAUAUGCCCGUAGGUUUAUUUUUAGACUUCUGUUUGUUUACCUGCGA ---(((((((..(((((((....----....-)))))))..)))))))...........(((((((..((...(((....)))...))..))))))). ( -22.60, z-score = -4.08, R) >consensus AAAUAAAGUUGUACACAGUUCUUUAUAUUAUUCCUAUGGUAUAUAUUUUGUAAUAUGCCAGUAGGUUUAUUUAUAGACUUCUGUUUACCGACGA____ .......((((...((((...((((((.....((((((((((((........)))))))).))))......))))))...))))....))))...... (-12.64 = -14.00 + 1.36)

| Location | 22,066,006 – 22,066,113 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.14 |
| Shannon entropy | 0.17299 |
| G+C content | 0.40929 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -23.24 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 22066006 107 - 27905053 GCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGUUUAAGGAAAAGUAGUUUCAUGGAUAUUUCCAUUUUCAUUUUUUC------GUCGGUAAACAGAA ((((((((((..((.....))..))))))))))............(((((((.((((((((.(....(((((....)))))...)))))))))------.....))))))).. ( -31.90, z-score = -2.82, R) >droSim1.chr3R 21909628 107 - 27517382 GCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUCGGGUAAGGAAAAGUAGUUUCAUGGAUAUUUCCAUUUUAAUUUUUUC------GUCGGUAAACAGAA ((((((((((..((.....))..))))))))))(..((..((.((..((((...((((.....))))(((((....)))))....))))..))------.))..))..).... ( -30.20, z-score = -2.11, R) >droSec1.super_13 845903 107 - 2104621 GCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGGGUAAGGAAAAGUAAUUCCAUGGAUAUUUCCAUUUUAAUUUUUUC------GUCGGUAAACAGAA ((((((((((..((.....))..))))))))))(..((..((.((..((((..((((......))))(((((....)))))....))))..))------.))..))..).... ( -33.90, z-score = -3.01, R) >droYak2.chr3R 4413565 113 + 28832112 GCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGGGUAAGGAAAAGUAGUUUCUUGGAUAUUUCCAUUUUCAUUUUUUUUUUUUGGUAGGUAAACAGAA ((((((((((..((.....))..))))))))))(..((......((..((.((((((((((.(.....((((....))))....)))))))))))))..))...))..).... ( -29.30, z-score = -1.46, R) >droEre2.scaffold_4820 4483743 111 + 10470090 GCCGGGCCUGAAAUGUUUAAUAGUGGGCUGGGCGGGUAAAGAGGAUUGGUUAGGGAAAAGUAGUUUUAUGGAUAUUUCCAUUUUCCUUUUGUCG--CAGGUAAACAAACAGAA (((.((((((..((.....))..)))))).)))(..((.....(((.....(((((((.........(((((....))))))))))))..))).--....))..)........ ( -26.60, z-score = -0.13, R) >consensus GCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGGGUAAGGAAAAGUAGUUUCAUGGAUAUUUCCAUUUUCAUUUUUUC______GUCGGUAAACAGAA ((((((((((..((.....))..))))))))))....................((((((((......(((((....)))))....)))))))).................... (-23.24 = -23.88 + 0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:45:05 2011