
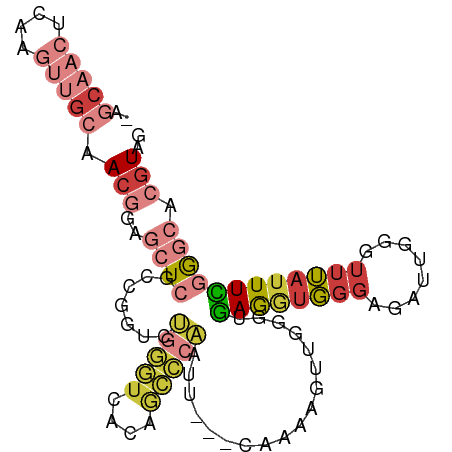
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,987,843 – 21,987,949 |
| Length | 106 |
| Max. P | 0.662140 |

| Location | 21,987,843 – 21,987,938 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 70.08 |
| Shannon entropy | 0.60461 |
| G+C content | 0.50511 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -10.03 |
| Energy contribution | -11.96 |
| Covariance contribution | 1.93 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21987843 95 - 27905053 -AGCAACUCAAGUUGCAACGGAGCUCACCGGUCUGGGUCACAGCCCAAUU---CAAAAGUUGGGUGAGGUGGGAGAUUGGUUUUAUUUCGGGCACGUAG -.(((((....))))).(((..((((((((((((...((((.(((((((.---.....)))))))...)))).))))))))........)))).))).. ( -35.20, z-score = -3.08, R) >droSim1.chr2L 16104023 95 + 22036055 -AGCAACUCAAGUUGCAACGGAGCUCGCCGGUCUGGGUCACAGCCCAAUU---CAAAAGUUGGGUGAGGUGGGAGAUUGGGUUUAUUUCGGGCACGUAG -.(((((....))))).(((..((((((((((((...((((.(((((((.---.....)))))))...)))).)))))))........))))).))).. ( -35.90, z-score = -2.89, R) >droSec1.super_13 773724 95 - 2104621 -AGCAACUCAAGUUGCAACGGAGCUCGCCGGUCUGGGUCACAGCCCAAUU---CGAAAGUUGGGUGAGGUGGGAGAUUGGGUUUAUUUCGGGCACGUAG -.(((((....))))).(((..((((((((((((...((((.(((((((.---.....)))))))...)))).)))))))........))))).))).. ( -35.90, z-score = -2.62, R) >droYak2.chr3R 4339020 95 + 28832112 -AGCAACUCAAGUUGCAACGGAGCUCGCCCGUCUGGGUCAGAGCCCAAUU---CAAAAGUUGGCUGAGGUGGGAGAUUGGGUUUAUUUCGGGCACGUAG -.(((((....))))).(((..((((((((....))))..((((((((((---((..((....))....))...)))))))))).....)))).))).. ( -31.60, z-score = -0.99, R) >droEre2.scaffold_4820 4408452 95 + 10470090 -AGCAACUCAAGUUGCAACGGAGCUCGCCGGUCUGGGUCACAGCCCAAUU---CAAAAGUUGGGCGAGGUGGGAGAUUGGGUUUAUUUCGGGCACGUAG -.(((((....))))).(((..((((((((((((...((((.(((((((.---.....)))))))...)))).)))))))........))))).))).. ( -37.50, z-score = -3.31, R) >droPer1.super_3 5574853 84 + 7375914 ---------------AAAAAAAUCUAGUCUGAAAACGCAAUGAAACAGAUGGCCACAACUGGUGGUAGACGGCAAAUUGGGUUUAUUUAGGUCACGUAG ---------------...........((((((((((.((((..........(((((.....))))).........)))).)))..)))))).)...... ( -15.51, z-score = -0.23, R) >droWil1.scaffold_181130 9304701 94 + 16660200 AAACCCCUCAACUUGCAACGGAAGUUUGAUGGCAGAUUGGCAAAUAUGG----UGGCAGUGGCAGGAAAUUGGGAAUUCAAUGGAAUUUCA-CAGGUAG ..(((......((((((((....))).........((((.((.......----)).)))).)))))....(((((((((....))))))).-))))).. ( -21.20, z-score = 0.29, R) >consensus _AGCAACUCAAGUUGCAACGGAGCUCGCCGGUCUGGGUCACAGCCCAAUU___CAAAAGUUGGGUGAGGUGGGAGAUUGGGUUUAUUUCGGGCACGUAG ..(((((....))))).(((..((((.......(((((....)))))..................((((((((........)))))))))))).))).. (-10.03 = -11.96 + 1.93)


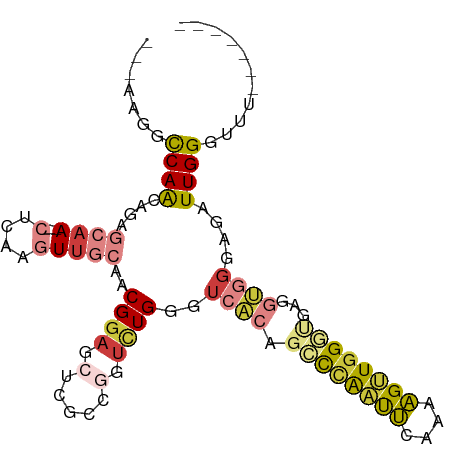
| Location | 21,987,858 – 21,987,949 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 69.84 |
| Shannon entropy | 0.55444 |
| G+C content | 0.51417 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -12.97 |
| Energy contribution | -13.07 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.511174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21987858 91 - 27905053 ---AAGGCCAACAGAGCAA---CUCAAGUUGCAACGGAGCUCACCGGUCUGGGUCACAGCCCAAUUCAAAAGUUGGGUGAGGUGGGAGAUUGGUUUU------- ---((((((((((((((((---(....)))))..(((......))).))))..((((.(((((((......)))))))...))))....))))))))------- ( -33.90, z-score = -2.50, R) >droSim1.chr2L 16104038 91 + 22036055 ---AAGGCCAACAGAGCAA---CUCAAGUUGCAACGGAGCUCGCCGGUCUGGGUCACAGCCCAAUUCAAAAGUUGGGUGAGGUGGGAGAUUGGGUUU------- ---(((.((((((((((((---(....)))))..(((......))).))))..((((.(((((((......)))))))...))))....)))).)))------- ( -32.10, z-score = -1.61, R) >droSec1.super_13 773739 91 - 2104621 ---AAGGCCAACAGAGCAA---CUCAAGUUGCAACGGAGCUCGCCGGUCUGGGUCACAGCCCAAUUCGAAAGUUGGGUGAGGUGGGAGAUUGGGUUU------- ---(((.((((((((((((---(....)))))..(((......))).))))..((((.(((((((......)))))))...))))....)))).)))------- ( -32.10, z-score = -1.42, R) >droYak2.chr3R 4339035 91 + 28832112 ---AAGGCCAACAGAGCAA---CUCAAGUUGCAACGGAGCUCGCCCGUCUGGGUCAGAGCCCAAUUCAAAAGUUGGCUGAGGUGGGAGAUUGGGUUU------- ---..(((((((...((((---(....)))))...((.((((((((....))))..)))))).........)))))))...................------- ( -32.40, z-score = -1.30, R) >droEre2.scaffold_4820 4408467 91 + 10470090 ---AAGGCCAACAGAGCAA---CUCAAGUUGCAACGGAGCUCGCCGGUCUGGGUCACAGCCCAAUUCAAAAGUUGGGCGAGGUGGGAGAUUGGGUUU------- ---(((.((((((((((((---(....)))))..(((......))).))))..((((.(((((((......)))))))...))))....)))).)))------- ( -33.70, z-score = -2.03, R) >droPer1.super_3 5574868 83 + 7375914 AAUGCCGUCAGCAGAAAAAAAUCUAGUCUGAAAACGCAA----------UGA--AACAGAUGGCCACAACUGGUGGUA--GACGGCAAAUUGGGUUU------- ..(((((((................(((((.........----------...--..))))).(((((.....))))).--)))))))..........------- ( -22.94, z-score = -2.19, R) >droWil1.scaffold_181130 9304715 93 + 16660200 ---AGGACCAAUACCCAAACCCCUCAACUUGCAACGGAA-------GUUUGA-UGGCAGAUUGGCAAAUAUGGUGGCAGUGGCAGGAAAUUGGGAAUUCAAUGG ---..........(((((...((((.(((.(((((....-------)))...-..((......))..........))))).).)))...))))).......... ( -18.20, z-score = 1.99, R) >consensus ___AAGGCCAACAGAGCAA___CUCAAGUUGCAACGGAGCUCGCCGGUCUGGGUCACAGCCCAAUUCAAAAGUUGGGUGAGGUGGGAGAUUGGGUUU_______ .......((((.......................((((.(.....).))))..((((.((((((((....))))))))...))))....))))........... (-12.97 = -13.07 + 0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:59 2011