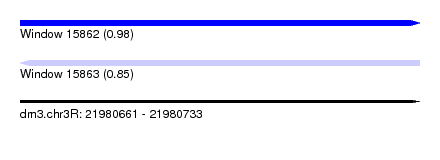
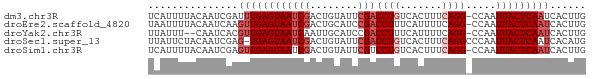
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,980,661 – 21,980,733 |
| Length | 72 |
| Max. P | 0.983353 |

| Location | 21,980,661 – 21,980,733 |
|---|---|
| Length | 72 |
| Sequences | 5 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 88.67 |
| Shannon entropy | 0.19614 |
| G+C content | 0.37698 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -14.12 |
| Energy contribution | -14.72 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
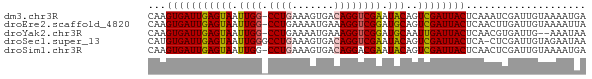
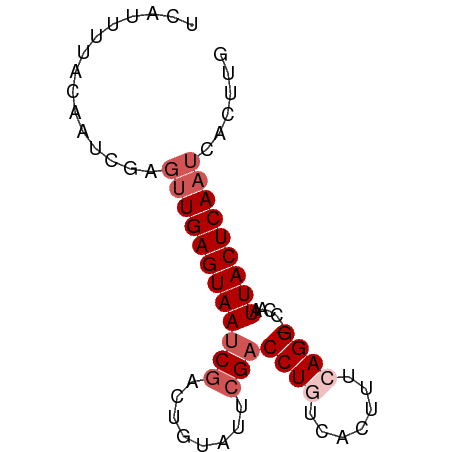
>dm3.chr3R 21980661 72 + 27905053 CAAGUGAUUGAGUAAUUGG-CCUGAAAGUGACAGGUCGAAUACAGUCGAUUACUCAAAUCGAUUGUAAAAUGA (((....))).....((((-((((.......)))))))).(((((((((((.....)))))))))))...... ( -23.40, z-score = -4.45, R) >droEre2.scaffold_4820 4401203 72 - 10470090 CAAGUGAUUGAGUAAUUGG-CCUGAAAAUGAAAGGUCGGAUGCAGUCGAUUACUCAACUUGAUUGUAAAAUUA (((((...(((((((((((-(.((....(((....)))....)))))))))))))))))))............ ( -21.10, z-score = -3.92, R) >droYak2.chr3R 4331945 70 - 28832112 CAAGUGAUUGAGUAAUUGG-CCUGAAAAUGAAAGGUCGGAUGCAAUUGAUUACUCAACGUGAUUG--AAAUAA (((....))).(((.((((-(((.........))))))).)))..(..(((((.....)))))..--)..... ( -16.00, z-score = -2.59, R) >droSec1.super_13 766577 72 + 2104621 CAUGUGAUUGAGUAAUUGGGCCUGAAAGUGACAGGUCGAAUACAGUCGAUUACUCA-CUCGAUUGUAGAAUAA ....(((.((((((((((((((((.......))))))(....)...))))))))))-.)))............ ( -20.10, z-score = -2.32, R) >droSim1.chr3R 21836971 72 + 27517382 CAAGUGAUUGAGUAAUUGG-CCUGAAAGUGACAGGACGAAUACAGUCGAUUACUCAACUCGAUUGUAAAAUGA (((.(((((((((((((((-(.((...((......)).....))))))))))))))).))).)))........ ( -18.40, z-score = -2.44, R) >consensus CAAGUGAUUGAGUAAUUGG_CCUGAAAGUGACAGGUCGAAUACAGUCGAUUACUCAACUCGAUUGUAAAAUAA (((.(((((((((((((((..(((....(((....)))....))))))))))))))).))).)))........ (-14.12 = -14.72 + 0.60)
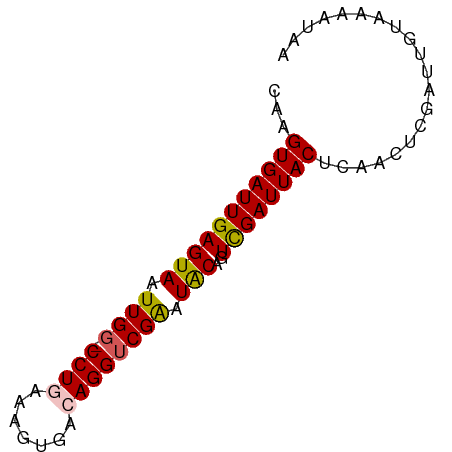
| Location | 21,980,661 – 21,980,733 |
|---|---|
| Length | 72 |
| Sequences | 5 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 88.67 |
| Shannon entropy | 0.19614 |
| G+C content | 0.37698 |
| Mean single sequence MFE | -13.30 |
| Consensus MFE | -9.08 |
| Energy contribution | -10.28 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21980661 72 - 27905053 UCAUUUUACAAUCGAUUUGAGUAAUCGACUGUAUUCGACCUGUCACUUUCAGG-CCAAUUACUCAAUCACUUG .............((((.((((((((((......))))((((.......))))-....))))))))))..... ( -15.70, z-score = -3.29, R) >droEre2.scaffold_4820 4401203 72 + 10470090 UAAUUUUACAAUCAAGUUGAGUAAUCGACUGCAUCCGACCUUUCAUUUUCAGG-CCAAUUACUCAAUCACUUG ............(((((((((((((.(.(((.....((....)).....))).-.).))))))))...))))) ( -12.30, z-score = -2.74, R) >droYak2.chr3R 4331945 70 + 28832112 UUAUUU--CAAUCACGUUGAGUAAUCAAUUGCAUCCGACCUUUCAUUUUCAGG-CCAAUUACUCAAUCACUUG ......--.......((((((((((...(((.....((....)).....))).-...))))))))))...... ( -8.50, z-score = -1.63, R) >droSec1.super_13 766577 72 - 2104621 UUAUUCUACAAUCGAG-UGAGUAAUCGACUGUAUUCGACCUGUCACUUUCAGGCCCAAUUACUCAAUCACAUG .............((.-(((((((((((......))))((((.......)))).....))))))).))..... ( -15.80, z-score = -2.62, R) >droSim1.chr3R 21836971 72 - 27517382 UCAUUUUACAAUCGAGUUGAGUAAUCGACUGUAUUCGUCCUGUCACUUUCAGG-CCAAUUACUCAAUCACUUG ............(((((((((((((.((......))(.((((.......))))-.).))))))))...))))) ( -14.20, z-score = -2.21, R) >consensus UCAUUUUACAAUCGAGUUGAGUAAUCGACUGUAUUCGACCUGUCACUUUCAGG_CCAAUUACUCAAUCACUUG ...............((((((((((((........)))((((.......)))).....)))))))))...... ( -9.08 = -10.28 + 1.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:56 2011