| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,977,325 – 21,977,452 |
| Length | 127 |
| Max. P | 0.626481 |

| Location | 21,977,325 – 21,977,422 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 93.68 |
| Shannon entropy | 0.10127 |
| G+C content | 0.41387 |
| Mean single sequence MFE | -8.37 |
| Consensus MFE | -8.43 |
| Energy contribution | -8.18 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.70 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
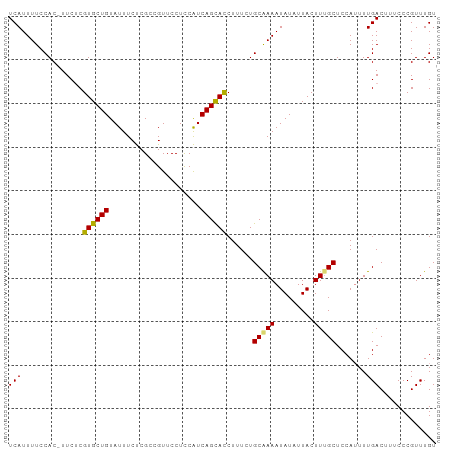
>dm3.chr3R 21977325 97 - 27905053 UCAUUUUCCAC-UUCUCGUGCUGUAUUUCUCGCCGUUCCUCCAUCAGUACCUUUUUGCAAAAUAUAUUACUUUGCUCCAUUUUGACUUCCCCGUUUGU (((........-.....((((((.....................))))))......(((((.........))))).......)))............. ( -8.00, z-score = -1.18, R) >droSim1.chr3R 21832714 97 - 27517382 UCAUUUUCCAC-UUCUCGUGCUGUAUUUCUCGCCGUUCCUCCAUCAGCACCUUUCUGCGAAAUAUAUUACUUUGCUCCAGUUUGACUUUCCCGUUUGU (((........-.....((((((.....................))))))......(((((.........))))).......)))............. ( -9.70, z-score = -0.49, R) >droSec1.super_13 763071 97 - 2104621 UCAUUUUCCAC-UUCUCGUGCUGUAUUUCUCGCCGUUCCUCCAUCAGCAUCUUUCUGCAAAAUAUAUUACUUUGCUCCAUUUUGACUUUCCCGUUUGU (((........-.....((((((.....................))))))......(((((.........))))).......)))............. ( -8.00, z-score = -0.82, R) >droEre2.scaffold_4820 4395976 98 + 10470090 UCAUUUUCCACUUUCACGUGCUGUAUUUCCCGCCGUUCCUUCGUCAGCACCUUUUUGCAAAAUAUAUUACUUCGCUCCAUUUUGACUUUCCCGUUUGU .................((((((.....................)))))).......((((((...............)))))).............. ( -7.76, z-score = -0.29, R) >consensus UCAUUUUCCAC_UUCUCGUGCUGUAUUUCUCGCCGUUCCUCCAUCAGCACCUUUCUGCAAAAUAUAUUACUUUGCUCCAUUUUGACUUUCCCGUUUGU (((..............((((((.....................))))))......(((((.........))))).......)))............. ( -8.43 = -8.18 + -0.25)


| Location | 21,977,351 – 21,977,452 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.91 |
| Shannon entropy | 0.27729 |
| G+C content | 0.42268 |
| Mean single sequence MFE | -12.56 |
| Consensus MFE | -11.13 |
| Energy contribution | -11.12 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.588178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
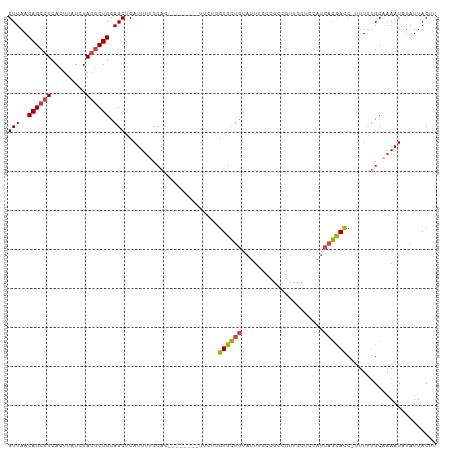
>dm3.chr3R 21977351 101 - 27905053 GUUAACAGCCUCACUUAUCUAGGCUGCAGCUCAUUUUCCAC--------UUCUCGUGCUGUAUUUCUCGCCGUUCCUCCAUCAGUACC-UUUUUGCAAAAUAUAUUACUU (((..((((((.........)))))).)))..(((((.((.--------.....((((((.....................)))))).-....)).)))))......... ( -13.40, z-score = -1.31, R) >droSim1.chr3R 21832740 101 - 27517382 GUUAACAGCCUCACUUAUCUAGGCUGCAGCUCAUUUUCCAC--------UUCUCGUGCUGUAUUUCUCGCCGUUCCUCCAUCAGCACC-UUUCUGCGAAAUAUAUUACUU (((..((((((.........)))))).)))...........--------.....(((.((((((((.....(......)....(((..-....))))))))))).))).. ( -16.50, z-score = -1.80, R) >droSec1.super_13 763097 101 - 2104621 GUUAACAGCCUCACUUAUCUAGGCUGCAGCUCAUUUUCCAC--------UUCUCGUGCUGUAUUUCUCGCCGUUCCUCCAUCAGCAUC-UUUCUGCAAAAUAUAUUACUU (((..((((((.........)))))).)))..(((((.((.--------.....((((((.....................)))))).-....)).)))))......... ( -13.90, z-score = -1.36, R) >droYak2.chr3R 4328450 110 + 28832112 GUUAACAGCCUCACUUAUCUAGACUGCAGCUCAUUUUCCACUUUCCACUUUCUCGUGCUGUAUUUCCCGCCGUUCCUUUAUCAGCACCUUUUUUGCAAAAUAUAUUACUU ...(((.((....((.....))..((((((..........................))))))......)).))).........(((.......))).............. ( -8.77, z-score = 0.05, R) >droEre2.scaffold_4820 4396002 102 + 10470090 GUUAACAGCCUCACUUAUCUAGGCUGCAGCUCAUUUUCCACU-------UUCACGUGCUGUAUUUCCCGCCGUUCCUUCGUCAGCACC-UUUUUGCAAAAUAUAUUACUU (((..((((((.........)))))).)))............-------.....((((((.....................)))))).-..................... ( -15.00, z-score = -1.28, R) >droAna3.scaffold_13340 12010260 91 + 23697760 GUUAACAGCAUCACUUAUCUAAGCUGCAGCUCAUUUUCCACU-------UGCUUGUGUU---CUUUCCCCCUUUCC-------ACACCUCCUUGGC--AAUUUAUUACUU (((((.((((...........(((....)))...........-------))))((((..---.............)-------))).....)))))--............ ( -7.81, z-score = 0.11, R) >consensus GUUAACAGCCUCACUUAUCUAGGCUGCAGCUCAUUUUCCAC________UUCUCGUGCUGUAUUUCCCGCCGUUCCUCCAUCAGCACC_UUUUUGCAAAAUAUAUUACUU (((..((((((.........)))))).)))........................((((((.....................))))))....................... (-11.13 = -11.12 + -0.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:55 2011