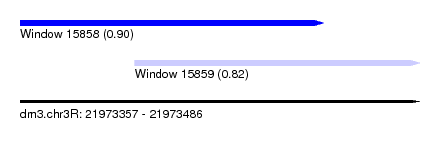
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,973,357 – 21,973,486 |
| Length | 129 |
| Max. P | 0.901589 |

| Location | 21,973,357 – 21,973,455 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.93 |
| Shannon entropy | 0.49979 |
| G+C content | 0.37792 |
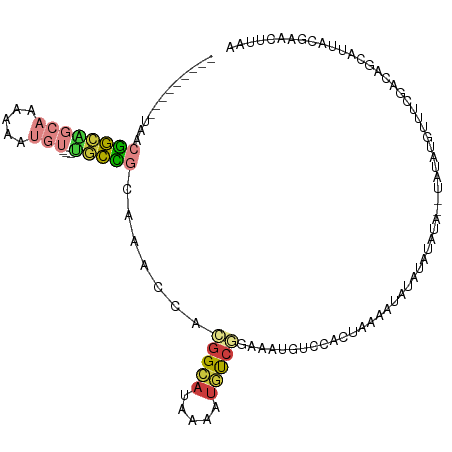
| Mean single sequence MFE | -22.73 |
| Consensus MFE | -13.44 |
| Energy contribution | -14.03 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
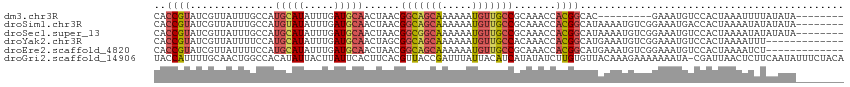
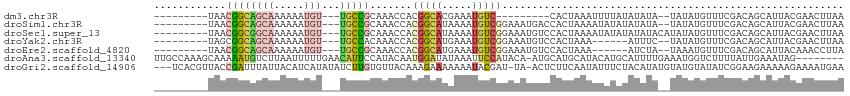
>dm3.chr3R 21973357 98 + 27905053 CACCGUAUCGUUAUUUGCCAUGCAUAUUUGAUGCAACUAACGGCAGCAAAAAAUGUUGCCGCAAACCACGGCAC---------GAAAUGUCCACUAAAUUUUAUAUA-------- ...(((..(((..(((((..(((((.....)))))......(((((((.....))))))))))))..)))..))---------).......................-------- ( -27.50, z-score = -3.12, R) >droSim1.chr3R 21829038 107 + 27517382 CACCGUAUCGUUAUUUGCCAUGUAUAUUUGAUGCAACUAACGGCAGCAAAAAAUGUUGCCGCAAACCACGGCAUAAAAUGUCGGAAAUGACCACUAAAAUAUAUAUA-------- ..(((...((((...((((.(((((.....))))).....((((((((.....))))))))........))))...)))).))).......................-------- ( -27.50, z-score = -2.51, R) >droSec1.super_13 759112 107 + 2104621 CACCGUAUCGUUAUUUGCCAUGCAUAUUUGAUGCAACUAACGGCGGCAAAAAAUGUUGCCGCAAACCACGGCAUAAAAUGUCGGAAAUGUCCACUAAAAUAUAUAUA-------- ..(((...((((...((((.(((((.....))))).......(((((((......))))))).......))))...)))).))).......................-------- ( -29.60, z-score = -2.42, R) >droYak2.chr3R 4324430 102 - 28832112 CACCGUAUCGUUAUUUUCCAUGCAUAUUUGAUGCAACUAGCGGCAGCAAAAAAUGUUGCCACAAACCACGGCAUGAAAUGUCGGAAAUGUCCACUAAAAUUU------------- ..............(((((.(((((.....)))))....(.(((((((.....))))))).).......(((((...))))))))))...............------------- ( -23.10, z-score = -1.01, R) >droEre2.scaffold_4820 4392163 102 - 10470090 CACCGUAUCGUUAUUUUCCAUGCAUAUUUGAUGCAACUAACGGCAGCAAAAAAUGUUGCCGCAAACCACGGCAUGAAAUGUCGGAAAUGUCCACUAAAAUCU------------- ..............(((((.(((((.....))))).....((((((((.....))))))))........(((((...))))))))))...............------------- ( -24.10, z-score = -1.49, R) >droGri2.scaffold_14906 10954651 114 + 14172833 UACCAUUUUGCAACUGGCCACAUAUUACUUAUUCACUUCACGUUACCGAUUUAUUACAUCAUAUAUCUUGUGUUACAAAGAAAAAAAUA-CGAUUAACUCUUCAAUAUUUCUACA ..(((.........)))........................((((..(((.(((......))).)))(((((((...........))))-))).))))................. ( -4.60, z-score = 1.68, R) >consensus CACCGUAUCGUUAUUUGCCAUGCAUAUUUGAUGCAACUAACGGCAGCAAAAAAUGUUGCCGCAAACCACGGCAUAAAAUGUCGGAAAUGUCCACUAAAAUUUAUAUA________ ..((((..............(((((.....)))))......(((((((.....))))))).......))))............................................ (-13.44 = -14.03 + 0.59)
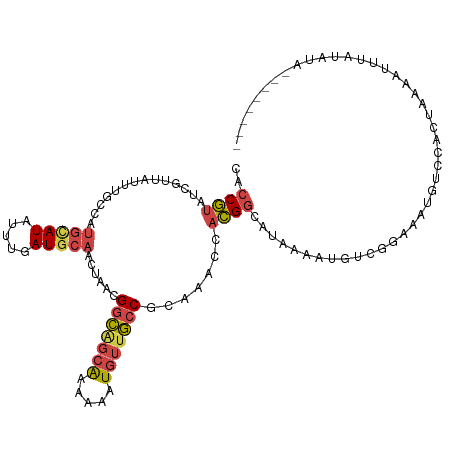
| Location | 21,973,394 – 21,973,486 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 58.81 |
| Shannon entropy | 0.79064 |
| G+C content | 0.34014 |
| Mean single sequence MFE | -20.69 |
| Consensus MFE | -5.34 |
| Energy contribution | -4.85 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.90 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21973394 92 + 27905053 ---------UAACGGCAGCAAAAAAUGU---UGCCGCAAACCACGGCACGAAAUGUC---------CACUAAAUUUUAUAUAUA--UAUAUGUUUCGACAGCAUUACGAACUUAA ---------...((((((((.....)))---)))))............((.(((((.---------..(.((((..((((....--)))).)))).)...))))).))....... ( -16.60, z-score = -1.06, R) >droSim1.chr3R 21829075 101 + 27517382 ---------UAACGGCAGCAAAAAAUGU---UGCCGCAAACCACGGCAUAAAAUGUCGGAAAUGACCACUAAAAUAUAUAUAUA--UAUAUGUUUCGACAGCAUUACGAACUUAA ---------...((((((((.....)))---))))).......((((((...))))))(.((((.......(((((((((....--)))))))))......)))).)........ ( -22.72, z-score = -2.76, R) >droSec1.super_13 759149 103 + 2104621 ---------UAACGGCGGCAAAAAAUGU---UGCCGCAAACCACGGCAUAAAAUGUCGGAAAUGUCCACUAAAAUAUAUAUAUACAUAUAUGUUUCGACAGCAUUACGAACUUAA ---------...(((((((((......)---))))))......((((((...))))))....((((.....((((((((((....)))))))))).))))......))....... ( -25.50, z-score = -2.87, R) >droYak2.chr3R 4324467 95 - 28832112 ---------UAGCGGCAGCAAAAAAUGU---UGCCACAAACCACGGCAUGAAAUGUCGGAAAUGUCCACUAAA------AUUUC--UAUAUGUUUCGACAGCAUUACGAACUUAA ---------..(.(((((((.....)))---)))).)......((..(((...((((((((((((........------.....--.)))).)))))))).)))..))....... ( -22.44, z-score = -2.05, R) >droEre2.scaffold_4820 4392200 95 - 10470090 ---------UAACGGCAGCAAAAAAUGU---UGCCGCAAACCACGGCAUGAAAUGUCGGAAAUGUCCACUAAA------AUCUA--UAAAUGUUUCGACAGCAUUACAAACCUUA ---------...((((((((.....)))---)))))........(..(((...((((((((............------.....--......)))))))).)))..)........ ( -20.60, z-score = -1.75, R) >droAna3.scaffold_13340 12030852 106 - 23697760 UUGCCAAAGCAAAAAUGUCUUAAUUUUUGAACAUUCCAUACAAUGGAUAUAAAUUCCAUACA-AUGCAUGCAUACAUGCAUUUUGAAAUGGUCUUUUAUUGAAAUAG-------- ((((....))))...(((.(((((....((.((((.((....(((((.......)))))..(-(((((((....)))))))).)).)))).))....))))).))).-------- ( -21.00, z-score = -1.48, R) >droGri2.scaffold_14906 10954688 110 + 14172833 ---UCACGUUACCGAUUUAUUACAUCAUAUAUCUUGUGUUACAAAGAAAAAAAUACGAU-UA-ACUCUUCAAUAUUUCUACAUAUGUAUGUAUAUCGGAAGAAAAAGAAAAUGAA ---........(((((....((((((((((.((((........)))).........((.-..-.....))...........))))).))))).)))))................. ( -16.00, z-score = -0.71, R) >consensus _________UAACGGCAGCAAAAAAUGU___UGCCGCAAACCACGGCAUAAAAUGUCGGAAAUGUCCACUAAAAUAUAUAUAUA__UAUAUGUUUCGACAGCAUUACGAACUUAA ............(((((..............))))).......(((((.....)))))......................................................... ( -5.34 = -4.85 + -0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:53 2011