
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,703,736 – 8,703,830 |
| Length | 94 |
| Max. P | 0.938086 |

| Location | 8,703,736 – 8,703,830 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.87 |
| Shannon entropy | 0.44478 |
| G+C content | 0.38361 |
| Mean single sequence MFE | -21.95 |
| Consensus MFE | -13.95 |
| Energy contribution | -13.95 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
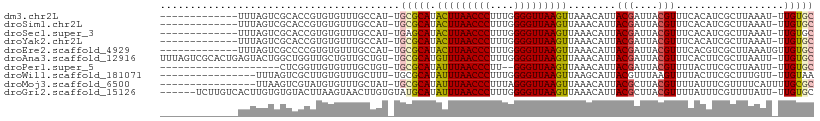
>dm3.chr2L 8703736 94 + 23011544 -------------UUUAGUCGCACCGUGUGUUUGCCAU-UGCGCAUACUUAACCCUUUGGGGUUAAGUUAAACAUUACGAUUACGUUUCACAUCGCUUAAAU-UUGUGC -------------..((((((....((((((.......-.))))))(((((((((....))))))))).........)))))).....((((..........-.)))). ( -23.80, z-score = -1.77, R) >droSim1.chr2L 8488950 94 + 22036055 -------------UUUAGUCGCACCGUGUGUUUGCCAU-UGCGCAUACUUAACCCUUUGGGGUUAAGUUAAACAUUACGAUUACGUUUCACAUCGCUUAAAU-UUGUGC -------------..((((((....((((((.......-.))))))(((((((((....))))))))).........)))))).....((((..........-.)))). ( -23.80, z-score = -1.77, R) >droSec1.super_3 4185415 94 + 7220098 -------------UUUAGUCGCACCGUGUGUUUGCCAU-UGAGCAUACUUAACCCUUUGGGGUUAAGUUAAACAUUACGAUUACGUUUCACAUCGCUUAAAU-UUGUGC -------------.......(((((((((((((((...-...))..(((((((((....))))))))).))))).))))....((........)).......-..)))) ( -22.80, z-score = -1.64, R) >droYak2.chr2L 11352412 94 + 22324452 -------------UUUAGUCGCACCGUGUGUUUGCCAU-UGCGCAUACUUAACCCUUUGGGGUUAAGUUAAACAUUACGAUUACGUUUCACAUCGCUUAAAU-UUGUGC -------------..((((((....((((((.......-.))))))(((((((((....))))))))).........)))))).....((((..........-.)))). ( -23.80, z-score = -1.77, R) >droEre2.scaffold_4929 9297653 95 + 26641161 -------------UUUAGUCGCCCCGUGUGUUUGCCAU-UGCGCAUACUUAACCCUUUGGGGUUAAGUUAAACAUUACGAUUACGUUUCACGUCGCUUAAAUGUUGUGC -------------..((((((....((((((.......-.))))))(((((((((....))))))))).........)))))).....((((..((......)))))). ( -23.90, z-score = -1.26, R) >droAna3.scaffold_12916 10349195 107 + 16180835 UUUAGUCGCACUGAGUACUGGCUGGUUGCUGUUGCUGU-UGCGCAUGUUUAACCCUUUGGGGUUAAGUUAAACAUUACGAUUACGUUUCACUUCGCUUAAUU-UUGUGC .......((((..((((..(((.....)))..))))..-.(((.....(((((((....)))))))((.((((...........)))).))..)))......-..)))) ( -23.10, z-score = 0.54, R) >droPer1.super_5 1451623 85 - 6813705 --------------------CUCGGUUGUGUUUGCUGU-UGCGCAUAUUUAACCCUU--GGGUUAAGUUAAACAUUACGAUUACGUUUUACUUCGCUUAAUU-UUGUGC --------------------.(((...((((((((...-.))....((((((((...--.)))))))).))))))..))).............(((......-..))). ( -17.10, z-score = -0.80, R) >droWil1.scaffold_181071 1144438 91 + 1211509 ----------------UUUAGUCGCUUGUGUUUGCUUU-UGCGCAUAUUUAACCCUUUGGGGUUAAGUUAAGCAUUACGUUUAAGUUUUACUUCGCUUUGUU-UUGUAA ----------------.......((((((((.......-.))))..(((((((((....))))))))).)))).(((((...((((........))))....-.))))) ( -19.70, z-score = -1.56, R) >droMoj3.scaffold_6500 10649719 92 + 32352404 ----------------UUAAGUCGUAUGUGUUUGCUAU-UGCGCAUAUUUAACCCUUUAGGGUUAAGUUAAACAUUACGCUUACGUUUUAUUUCGUUUUCAUUUUGCGC ----------------.((((.((((.((((((((...-.))....(((((((((....))))))))).)))))))))))))).......................... ( -21.40, z-score = -2.50, R) >droGri2.scaffold_15126 3634022 102 + 8399593 ------UCUUGUCACUUGUGUGUACUUAAGUAACUUGUGUAUGCAUAUUUAACCCUUUGGGGUUAAGUUAAACAUUACGCUUACGUUUUAUUUCGUUUUAUU-UUGUGC ------...............((((..((((((...(((((((...(((((((((....)))))))))....)).)))))..(((........))).)))))-).)))) ( -20.10, z-score = -1.29, R) >consensus _____________UUUAGUCGCACCGUGUGUUUGCCAU_UGCGCAUACUUAACCCUUUGGGGUUAAGUUAAACAUUACGAUUACGUUUCACUUCGCUUAAAU_UUGUGC ........................................(((((.(((((((((....)))))))))........(((....)))..................))))) (-13.95 = -13.95 + 0.00)
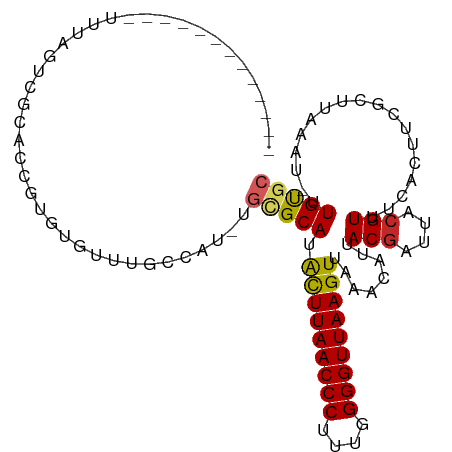
| Location | 8,703,736 – 8,703,830 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.87 |
| Shannon entropy | 0.44478 |
| G+C content | 0.38361 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -11.47 |
| Energy contribution | -12.01 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8703736 94 - 23011544 GCACAA-AUUUAAGCGAUGUGAAACGUAAUCGUAAUGUUUAACUUAACCCCAAAGGGUUAAGUAUGCGCA-AUGGCAAACACACGGUGCGACUAAA------------- ((((..-......(((((..........)))))..(((((.(((((((((....)))))))))..((...-...)))))))....)))).......------------- ( -26.00, z-score = -2.23, R) >droSim1.chr2L 8488950 94 - 22036055 GCACAA-AUUUAAGCGAUGUGAAACGUAAUCGUAAUGUUUAACUUAACCCCAAAGGGUUAAGUAUGCGCA-AUGGCAAACACACGGUGCGACUAAA------------- ((((..-......(((((..........)))))..(((((.(((((((((....)))))))))..((...-...)))))))....)))).......------------- ( -26.00, z-score = -2.23, R) >droSec1.super_3 4185415 94 - 7220098 GCACAA-AUUUAAGCGAUGUGAAACGUAAUCGUAAUGUUUAACUUAACCCCAAAGGGUUAAGUAUGCUCA-AUGGCAAACACACGGUGCGACUAAA------------- ((((..-......(((((..........)))))..(((((.(((((((((....)))))))))..(((..-..))))))))....)))).......------------- ( -26.30, z-score = -2.87, R) >droYak2.chr2L 11352412 94 - 22324452 GCACAA-AUUUAAGCGAUGUGAAACGUAAUCGUAAUGUUUAACUUAACCCCAAAGGGUUAAGUAUGCGCA-AUGGCAAACACACGGUGCGACUAAA------------- ((((..-......(((((..........)))))..(((((.(((((((((....)))))))))..((...-...)))))))....)))).......------------- ( -26.00, z-score = -2.23, R) >droEre2.scaffold_4929 9297653 95 - 26641161 GCACAACAUUUAAGCGACGUGAAACGUAAUCGUAAUGUUUAACUUAACCCCAAAGGGUUAAGUAUGCGCA-AUGGCAAACACACGGGGCGACUAAA------------- .............((..((((..(((....)))..(((((.(((((((((....)))))))))..((...-...)))))))))))..)).......------------- ( -26.60, z-score = -2.30, R) >droAna3.scaffold_12916 10349195 107 - 16180835 GCACAA-AAUUAAGCGAAGUGAAACGUAAUCGUAAUGUUUAACUUAACCCCAAAGGGUUAAACAUGCGCA-ACAGCAACAGCAACCAGCCAGUACUCAGUGCGACUAAA ((((..-......(((..(((((((((.......))))))...(((((((....))))))).))).))).-...((....))................))))....... ( -23.30, z-score = -1.48, R) >droPer1.super_5 1451623 85 + 6813705 GCACAA-AAUUAAGCGAAGUAAAACGUAAUCGUAAUGUUUAACUUAACCC--AAGGGUUAAAUAUGCGCA-ACAGCAAACACAACCGAG-------------------- ......-......(((..((.((((((.......)))))).))((((((.--...)))))).....))).-..................-------------------- ( -14.00, z-score = -1.09, R) >droWil1.scaffold_181071 1144438 91 - 1211509 UUACAA-AACAAAGCGAAGUAAAACUUAAACGUAAUGCUUAACUUAACCCCAAAGGGUUAAAUAUGCGCA-AAAGCAAACACAAGCGACUAAA---------------- ......-......((((((.....)))...)))..(((((...(((((((....)))))))...(((...-...))).....)))))......---------------- ( -15.90, z-score = -1.94, R) >droMoj3.scaffold_6500 10649719 92 - 32352404 GCGCAAAAUGAAAACGAAAUAAAACGUAAGCGUAAUGUUUAACUUAACCCUAAAGGGUUAAAUAUGCGCA-AUAGCAAACACAUACGACUUAA---------------- (((((...........(((((..(((....)))..)))))...(((((((....)))))))...))))).-......................---------------- ( -20.60, z-score = -3.10, R) >droGri2.scaffold_15126 3634022 102 - 8399593 GCACAA-AAUAAAACGAAAUAAAACGUAAGCGUAAUGUUUAACUUAACCCCAAAGGGUUAAAUAUGCAUACACAAGUUACUUAAGUACACACAAGUGACAAGA------ (((...-.........(((((..(((....)))..)))))...(((((((....)))))))...)))........(((((((..(....)..)))))))....------ ( -19.70, z-score = -2.68, R) >consensus GCACAA_AUUUAAGCGAAGUGAAACGUAAUCGUAAUGUUUAACUUAACCCCAAAGGGUUAAAUAUGCGCA_AUAGCAAACACACGGGGCGACUAAA_____________ .............(((..((.((((((.......)))))).))(((((((....))))))).....)))........................................ (-11.47 = -12.01 + 0.54)

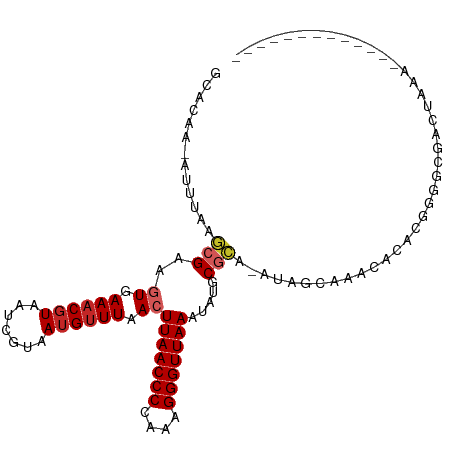
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:41 2011