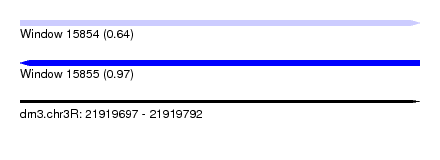
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,919,697 – 21,919,792 |
| Length | 95 |
| Max. P | 0.966377 |

| Location | 21,919,697 – 21,919,792 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.74 |
| Shannon entropy | 0.47707 |
| G+C content | 0.50421 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -12.01 |
| Energy contribution | -12.32 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.641096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
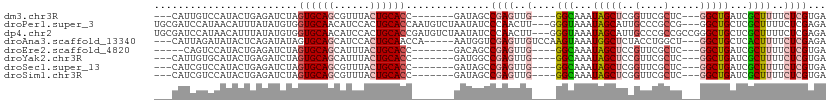
>dm3.chr3R 21919697 95 + 27905053 ---CAUUGUCCAUACUGAGAUCUAGUGCAGCGUUUACUGCACC-------GAUAGCCGAGUUG----GGCAAAUAGCUCGGUUCGCUC---GGCUGAUCGCUUUUCUCGUGA ---.........(((.((((....((((((......))))))(-------(((((((((((((----(((.....)))))....))))---)))).))))....))))))). ( -35.60, z-score = -2.58, R) >droPer1.super_3 5513841 106 - 7375914 UGCGAUCCAUAACAUUUAUAUGUGGUGCAACAUCCACUGCACCAAUGUCUAAUAUCCCAACUU---GGGUAAAUAGCAUUGCCCGCCG---GGCUGCUCGCUUUUCUCGAGA .((((......((((....))))((((((........))))))((((.(((....(((.....---)))....)))))))((((...)---)))...))))........... ( -28.60, z-score = -1.36, R) >dp4.chr2 11677302 109 - 30794189 UGCGAUCCAUAACAUUUAUAUGUGGUGCAACAUCCACUGCACCGAUGUCUAAUAUCCCAACUU---GGGUAAAUAGCAUUGCCCGCCGCCGGGCUGCUCGCUUUUCUCGAGA .((((......((((....))))((((((........))))))((((.(((....(((.....---)))....)))))))(((((....)))))...))))........... ( -32.20, z-score = -1.73, R) >droAna3.scaffold_13340 15746438 101 + 23697760 ---CAUUAGAUAUACUCAGAUAUAGUGCAGCAUCCACUGCAACCA-----AAUGGUCGAGUUGUCCAAGUAAAUGGCUCUACCUGGCU---GGCUGCUCACUUUUCUCGAGA ---...........((((((...((((.((((.(((..((.....-----...(((.((((..(........)..)))).)))..)))---)).))))))))..))).))). ( -28.40, z-score = -1.39, R) >droEre2.scaffold_4820 4346462 93 - 10470090 -----CAGUCCAUACUGAGAUCUAGUGCAGCAUUUACUGCACC-------GACAGCCGAGUUG----GGCAAAUAGCUCCGUUCGCUC---GGCUGAUCGCUUUUCUCGUGA -----.......(((.((((....((((((......))))))(-------(((((((((((((----(((.....)).)))...))))---))))).)))....))))))). ( -37.10, z-score = -3.53, R) >droYak2.chr3R 4277972 95 - 28832112 ---CAUUGUGCAUACUGAGAUCUAGUGCAGCAUUUACUGCACC-------GAUGGCCGAGUUG----GGCAAAUAGCUCCGUUCGCUC---GGCUGAUCGCUUUUCUCGUGA ---.........(((.((((....((((((......))))))(-------(((((((((((((----(((.....)).)))...))))---)))).))))....))))))). ( -34.90, z-score = -2.25, R) >droSec1.super_13 714212 95 + 2104621 ---CAUCGUCCAUACUGAGAUCUAGUGCAGCGUUUACUGCACC-------GAUAGCCGAGUUG----GGCAAAUAGCUCGGUUCGCUC---GGCUGAUCGCUUUUCUCGUGA ---.........(((.((((....((((((......))))))(-------(((((((((((((----(((.....)))))....))))---)))).))))....))))))). ( -35.60, z-score = -2.41, R) >droSim1.chr3R 21780281 95 + 27517382 ---CAUCGUCCAUACUGAGAUCUAGUGCAGCGUUUACUGCACC-------GAUAGCCGAGUUG----GGCAAAUAGCUCGGUUCGCUC---GGCUGAUCGCUUUUCUCGUGA ---.........(((.((((....((((((......))))))(-------(((((((((((((----(((.....)))))....))))---)))).))))....))))))). ( -35.60, z-score = -2.41, R) >consensus ___CAUCGUCCAUACUGAGAUCUAGUGCAGCAUUUACUGCACC_______GAUAGCCGAGUUG____GGCAAAUAGCUCGGUUCGCUC___GGCUGAUCGCUUUUCUCGUGA ........................((((((......))))))..............((((.......(((...((((...............))))...)))...))))... (-12.01 = -12.32 + 0.31)

| Location | 21,919,697 – 21,919,792 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.74 |
| Shannon entropy | 0.47707 |
| G+C content | 0.50421 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -16.89 |
| Energy contribution | -16.38 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
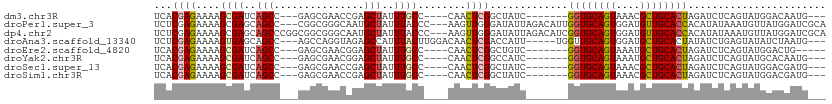
>dm3.chr3R 21919697 95 - 27905053 UCACGAGAAAAGCGAUCAGCC---GAGCGAACCGAGCUAUUUGCC----CAACUCGGCUAUC-------GGUGCAGUAAACGCUGCACUAGAUCUCAGUAUGGACAAUG--- ....((((....((((.((((---(((......(.((.....)))----...))))))))))-------)(((((((....)))))))....)))).............--- ( -31.20, z-score = -2.50, R) >droPer1.super_3 5513841 106 + 7375914 UCUCGAGAAAAGCGAGCAGCC---CGGCGGGCAAUGCUAUUUACCC---AAGUUGGGAUAUUAGACAUUGGUGCAGUGGAUGUUGCACCACAUAUAAAUGUUAUGGAUCGCA ...........((((.(.(((---(...))))(((((((....(((---.....)))....))).))))(((((((......))))))).((((.......))))).)))). ( -31.50, z-score = -0.99, R) >dp4.chr2 11677302 109 + 30794189 UCUCGAGAAAAGCGAGCAGCCCGGCGGCGGGCAAUGCUAUUUACCC---AAGUUGGGAUAUUAGACAUCGGUGCAGUGGAUGUUGCACCACAUAUAAAUGUUAUGGAUCGCA ...........((((.(.(((((....))))).((((((....(((---.....)))....))).))).(((((((......))))))).((((.......))))).)))). ( -34.20, z-score = -1.02, R) >droAna3.scaffold_13340 15746438 101 - 23697760 UCUCGAGAAAAGUGAGCAGCC---AGCCAGGUAGAGCCAUUUACUUGGACAACUCGACCAUU-----UGGUUGCAGUGGAUGCUGCACUAUAUCUGAGUAUAUCUAAUG--- .(((.(((..((((((((.((---.((((((((((....))))))))).).((((((((...-----.))))).))))).)))).))))...))))))...........--- ( -34.50, z-score = -2.97, R) >droEre2.scaffold_4820 4346462 93 + 10470090 UCACGAGAAAAGCGAUCAGCC---GAGCGAACGGAGCUAUUUGCC----CAACUCGGCUGUC-------GGUGCAGUAAAUGCUGCACUAGAUCUCAGUAUGGACUG----- ....((((....(((.(((((---(((.....((.((.....)))----)..))))))))))-------)(((((((....)))))))....))))...........----- ( -36.80, z-score = -3.44, R) >droYak2.chr3R 4277972 95 + 28832112 UCACGAGAAAAGCGAUCAGCC---GAGCGAACGGAGCUAUUUGCC----CAACUCGGCCAUC-------GGUGCAGUAAAUGCUGCACUAGAUCUCAGUAUGCACAAUG--- ....((((....((((..(((---(((.....((.((.....)))----)..)))))).)))-------)(((((((....)))))))....)))).............--- ( -32.30, z-score = -2.66, R) >droSec1.super_13 714212 95 - 2104621 UCACGAGAAAAGCGAUCAGCC---GAGCGAACCGAGCUAUUUGCC----CAACUCGGCUAUC-------GGUGCAGUAAACGCUGCACUAGAUCUCAGUAUGGACGAUG--- ....((((....((((.((((---(((......(.((.....)))----...))))))))))-------)(((((((....)))))))....)))).............--- ( -31.20, z-score = -2.12, R) >droSim1.chr3R 21780281 95 - 27517382 UCACGAGAAAAGCGAUCAGCC---GAGCGAACCGAGCUAUUUGCC----CAACUCGGCUAUC-------GGUGCAGUAAACGCUGCACUAGAUCUCAGUAUGGACGAUG--- ....((((....((((.((((---(((......(.((.....)))----...))))))))))-------)(((((((....)))))))....)))).............--- ( -31.20, z-score = -2.12, R) >consensus UCACGAGAAAAGCGAUCAGCC___GAGCGAACAGAGCUAUUUGCC____CAACUCGGCUAUC_______GGUGCAGUAAAUGCUGCACUAGAUCUCAGUAUGGACGAUG___ ...((((....((.....)).....(((.......)))..............)))).............((((((((....))))))))....................... (-16.89 = -16.38 + -0.51)
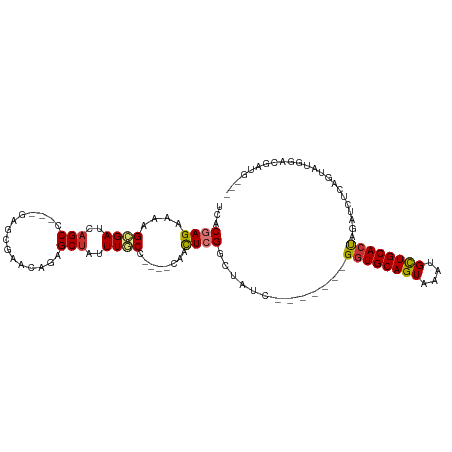
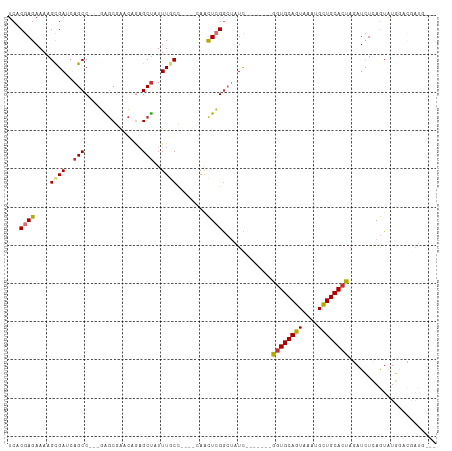
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:50 2011