
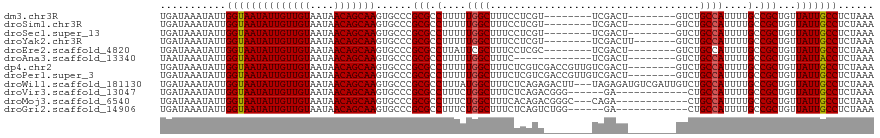
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,915,151 – 21,915,291 |
| Length | 140 |
| Max. P | 0.841187 |

| Location | 21,915,151 – 21,915,254 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.02 |
| Shannon entropy | 0.25324 |
| G+C content | 0.42049 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -20.46 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.841187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21915151 103 + 27905053 UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUUUGGCUUUCCUCGU--------UCGACU--------GUCUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ...........((((((((((((((....)))))))......(((.(....((((.....((..--------..))..--------....))))....).)))...)))))))...... ( -22.70, z-score = -1.38, R) >droSim1.chr3R 21775668 103 + 27517382 UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUUUGGCUUUCCUCGU--------UCGACU--------GUCUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ...........((((((((((((((....)))))))......(((.(....((((.....((..--------..))..--------....))))....).)))...)))))))...... ( -22.70, z-score = -1.38, R) >droSec1.super_13 709727 103 + 2104621 UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUUUGGCUUUCCUCGU--------UCGACU--------GUCUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ...........((((((((((((((....)))))))......(((.(....((((.....((..--------..))..--------....))))....).)))...)))))))...... ( -22.70, z-score = -1.38, R) >droYak2.chr3R 4273230 104 - 28832112 UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUUUGGCUUUCCUCGU--------UCGACUU-------GUCUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ...........(((((((..((.((((...(((((((((..((.(((.....)))......)).--------.).))))-------).))).....)))).))...)))))))...... ( -22.40, z-score = -1.39, R) >droEre2.scaffold_4820 4341820 103 - 10470090 UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUAUUCGCUUUCCUCGC--------UCGACU--------GUCUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ....................(..(((((((((((..(((....))).......((.......((--------......--------....))......)).)))))))))))..).... ( -18.42, z-score = -0.15, R) >droAna3.scaffold_13340 15741694 98 + 23697760 UAAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUUUGGCUUUC-------------UCGACU--------GUCUGCCAUUUUGCCGCUGUUAUUACCUCUAAA ...........((((((((((((((....)))))))......(((.(....((((...(-------------......--------)...))))....).)))...)))))))...... ( -23.00, z-score = -2.40, R) >dp4.chr2 11672325 111 - 30794189 UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUUUGGCUUUCUCGUCGACCGUUGUCGACU--------GUCUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ...........((((((((((((((....)))))))......(((.(....((((...(..((((((....)))))).--------)...))))....).)))...)))))))...... ( -31.50, z-score = -3.48, R) >droPer1.super_3 5508852 111 - 7375914 UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUUUGGCUUUCUCGUCGACCGUUGUCGACU--------GUCUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ...........((((((((((((((....)))))))......(((.(....((((...(..((((((....)))))).--------)...))))....).)))...)))))))...... ( -31.50, z-score = -3.48, R) >droWil1.scaffold_181130 15554214 116 + 16660200 UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUAUGGCUUUCUCAGAGACUU---UAGAGAUGUCGAUUGUCUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ...........((((((((((((((....)))))))......(((.(...(((((.....(((.(((..---.......)))..)))...)))))...).)))...)))))))...... ( -27.60, z-score = -1.28, R) >droVir3.scaffold_13047 5503467 101 + 19223366 UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUCUGGCUUUCUCAGACGGG------GA------------CUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ....................(..(((((((((((.(((.((((((((.....))).......).))))------.)------------))((......)).)))))))))))..).... ( -28.41, z-score = -1.62, R) >droMoj3.scaffold_6540 15878898 104 + 34148556 UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUCUGGCUUUCACAGACGGGC---CAGA------------CUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ....................(..(((((((((((..(((....)))...((((((((........))))---))))------------.............)))))))))))..).... ( -28.20, z-score = -1.19, R) >droGri2.scaffold_14906 10875139 101 + 14172833 UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUCUGGCUUUCUCAGUCUGG------GA------------CUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ....................(..(((((((((((..(((....))).....((((..((((.....))------))------------..)))).......)))))))))))..).... ( -27.90, z-score = -2.06, R) >consensus UGAUAAAUAUUGGUAAUAUUGUUGUAAUAACAGCAAGUGCCCGCGCCUUUUUGGCUUUCCCCGU________UCGACU________GUCUGCCAUUUUGCCGCUGUUAUUGCCUCUAAA ...........((((((((((((((....)))))))......(((.(....((((...................................))))....).)))...)))))))...... (-20.46 = -20.47 + 0.01)
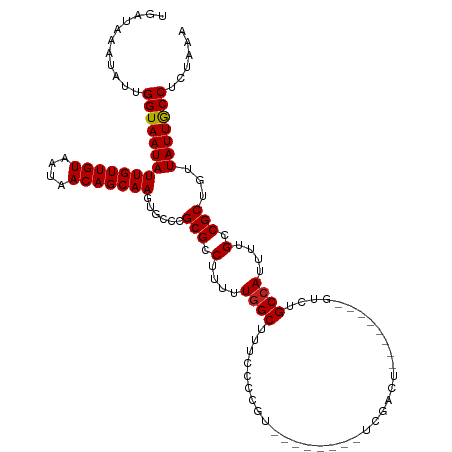

| Location | 21,915,191 – 21,915,291 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.33 |
| Shannon entropy | 0.29101 |
| G+C content | 0.46073 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -16.76 |
| Energy contribution | -16.77 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.769880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21915191 100 - 27905053 GGACGCA---GUGAGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAGAC--------AGUCG--AACG------AGGAAAGCCAAAAAGGCGCGG ...(((.---((..((((((((...(((....))).(((((((.(..((.......))..).)))))))))))--------)))).--.)).------......(((.....)))))). ( -31.00, z-score = -3.94, R) >droSim1.chr3R 21775708 100 - 27517382 GGACGCA---GUGAGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAGAC--------AGUCG--AACG------AGGAAAGCCAAAAAGGCGCGG ...(((.---((..((((((((...(((....))).(((((((.(..((.......))..).)))))))))))--------)))).--.)).------......(((.....)))))). ( -31.00, z-score = -3.94, R) >droSec1.super_13 709767 100 - 2104621 GGACGCA---GUGAGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAGAC--------AGUCG--AACG------AGGAAAGCCAAAAAGGCGCGG ...(((.---((..((((((((...(((....))).(((((((.(..((.......))..).)))))))))))--------)))).--.)).------......(((.....)))))). ( -31.00, z-score = -3.94, R) >droYak2.chr3R 4273270 101 + 28832112 GGACGCA---GUGAGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAGAC-------AAGUCG--AACG------AGGAAAGCCAAAAAGGCGCGG ...(((.---((..((((((((...(((....))).(((((((.(..((.......))..).)))))))))))-------.)))).--.)).------......(((.....)))))). ( -27.20, z-score = -2.51, R) >droEre2.scaffold_4820 4341860 100 + 10470090 GGACGCA---GUGAGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAGAC--------AGUCG--AGCG------AGGAAAGCGAAUAAGGCGCGG ...(((.---((..((((((((...(((....))).(((((((.(..((.......))..).)))))))))))--------)))).--.)).------......((.......))))). ( -27.80, z-score = -2.71, R) >droAna3.scaffold_13340 15741734 95 - 23697760 GGACGCA---GUGAGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGUAAUAACAGCGGCAAAAUGGCAGAC--------AGUC-------------GAGAAAGCCAAAAAGGCGCGG ...(((.---....((((((((...(((....))).(((((((.(..((.......))..).)))))))))))--------))))-------------......(((.....)))))). ( -26.90, z-score = -3.09, R) >dp4.chr2 11672365 108 + 30794189 GGACACA---GUGAGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAGAC--------AGUCGACAACGGUCGACGAGAAAGCCAAAAAGGCGCGG .......---((...(((((((((((((..........)))))))).))))).)).(((.(....((((....--------.((((((....))))))......))))....).))).. ( -29.30, z-score = -2.64, R) >droPer1.super_3 5508892 108 + 7375914 GGACACA---GUGAGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAGAC--------AGUCGACAACGGUCGACGAGAAAGCCAAAAAGGCGCGG .......---((...(((((((((((((..........)))))))).))))).)).(((.(....((((....--------.((((((....))))))......))))....).))).. ( -29.30, z-score = -2.64, R) >droWil1.scaffold_181130 15554254 116 - 16660200 GAAUACAUGUGUGUGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAGACAAUCGACAUCUCUAAAGUCUC---UGAGAAAGCCAUAAAGGCGCGG .........(((.(((((((((...(((....))).(((((((.(..((.......))..).)))))))))))))))))))((((.........---.))))..(((.....))).... ( -28.40, z-score = -1.04, R) >droVir3.scaffold_13047 5503507 101 - 19223366 AGAUACAUGUGUGUGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAG------------UC---CCCGUC---UGAGAAAGCCAGAAAGGCGCGG .....(((((.....(((......)))....)))))(((((((.(..((.......))..).))))))).(------------(.---((..((---((.......))))..)).)).. ( -23.70, z-score = 0.13, R) >droMoj3.scaffold_6540 15878938 104 - 34148556 AGAUACAUGUGUGUGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAG------------UCUGGCCCGUC---UGUGAAAGCCAGAAAGGCGCGG ((((.(((((.....(((......)))....)))))(((((((.(..((.......))..).))))))).)------------))).(((..((---(((....).))))..))).... ( -27.80, z-score = -0.25, R) >droGri2.scaffold_14906 10875179 101 - 14172833 AGAUACAUGUGUGUGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAG------------UC---CCAGAC---UGAGAAAGCCAGAAAGGCGCGG ...........(((.(((((((((((((..........)))))))).))))).)))(((.(....((((..------------((---.(....---.).))..))))....).))).. ( -23.40, z-score = -0.19, R) >consensus GGACACA___GUGAGAUUGUUUUAAAUGUUAACAUGGCCAUUUAGAGGCAAUAACAGCGGCAAAAUGGCAGAC________AGUCG__AACG______GAGAAAGCCAAAAAGGCGCGG ..........((...(((((((((((((..........)))))))).))))).)).(((.(....((((...................................))))....).))).. (-16.76 = -16.77 + 0.01)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:44 2011