
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,894,817 – 21,894,916 |
| Length | 99 |
| Max. P | 0.832642 |

| Location | 21,894,817 – 21,894,916 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.65 |
| Shannon entropy | 0.25217 |
| G+C content | 0.40607 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -15.59 |
| Energy contribution | -14.98 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.832642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21894817 99 - 27905053 UUUCAAUGAUUAUUCAUUGGGCAUGGCAGCCGCAAGC------CCGCACACAAG--------------CACACUUGUCUGUGGCAAACAAUUUCAGCAAUUAGAAUAUCAAGCUAUGCA ..(((((((....)))))))(((((((.(((....).------(((((.(((((--------------....))))).)))))............)).....(.....)..))))))). ( -29.70, z-score = -2.50, R) >droSim1.chr3R 21746242 99 - 27517382 UUUCAAUGAUUAUUCAUUGGGCAUGGCUGCCACAAGC------CCGCACACAAG--------------CACACUUGUCUGUGGCAAACAAUUUCAGCAAUUAGAAUAUCAAGCUAUGCA ..(((((((....)))))))((((((((((((((.(.------...)..(((((--------------....))))).)))))).......(((........))).....)))))))). ( -30.40, z-score = -2.85, R) >droSec1.super_13 689278 99 - 2104621 UUUCAAUGAUUAUUCAUUGGGCAUGGCUGCCACAAGC------CCGCACACAAG--------------CACACUUGUCUGCGGCAAACAAUUUCAGCAAUUAGAAUAUCAAGCUAUGCA ..(((((((....)))))))((((((((.......((------(.(((.(((((--------------....))))).)))))).......(((........))).....)))))))). ( -32.20, z-score = -3.72, R) >droYak2.chr3R 4252525 99 + 28832112 UUUCAAUGAUUAUUCAUUGGGCAUGGCAGCCACAAGC------CCGCACACAAG--------------CACACUUGUCUGUGGCAAACAAUUUCAGCAAUUAGAAUAUCAAGCUAUGCA ..(((((((....)))))))(((((((.((((((.(.------...)..(((((--------------....))))).)))))).......(((........)))......))))))). ( -29.90, z-score = -3.04, R) >droEre2.scaffold_4820 4320402 99 + 10470090 UUUCAAUGAUUAUUCAUUGGGCAUGGCAGCCACAAGC------CCGCACACAAG--------------CACACUUGUCUGUGGCAAACAAUUUCAGCAAUUAGAAUAUCAAGCUAUGCA ..(((((((....)))))))(((((((.((((((.(.------...)..(((((--------------....))))).)))))).......(((........)))......))))))). ( -29.90, z-score = -3.04, R) >droAna3.scaffold_13340 15720011 119 - 23697760 UUUCGAUGAUUAUUCAUUGGGCAUGGCAGUCACAAACACACAAUCACACACAGGCACAUACACAACGACACACUUGUCUGUGGCAAACAAUUUCAGCAAUUAGAAUAUCAAGCUAUGCA ..(((((((....)))))))(((((((.....................((((((((..................))))))))((...........))..............))))))). ( -27.37, z-score = -3.22, R) >droVir3.scaffold_13047 5479315 91 - 19223366 UUUCAAUGAUUAUUCAUUGGGCAAGGCGCAUAUAA--------UCGCA--------------------CACACUUGUCUGCGACAAACAAUUUCAGCAAUUAGAAUAUCAAGCAAUGCA ..(((((((....)))))))(((..((........--------(((((--------------------.((....)).)))))........(((........)))......))..))). ( -19.80, z-score = -1.96, R) >droMoj3.scaffold_6540 15853317 91 - 34148556 UUUCAAUGAUUAUUCAUUGGGCAUGGCGCAUAUAA--------UCGCA--------------------CACACUUGUCUGCGACAAACAAUUUCAGCAAUUAGAAUAUCAAGCAAUGCA ..(((((((....)))))))((((.((........--------(((((--------------------.((....)).)))))........(((........)))......)).)))). ( -20.80, z-score = -1.93, R) >droGri2.scaffold_14906 10853088 91 - 14172833 UUUCAAUGAUUAUUCAUUGGGCAUGGCGCAUAUAA--------UCACA--------------------CACACUUGUCUGCGACAAACAAUUUCAGCAAUUAGAAUAUCAAGCAAUGCA ..(((((((....)))))))((((.(((((.((((--------.....--------------------.....)))).)))..........(((........)))......)).)))). ( -16.40, z-score = -0.79, R) >consensus UUUCAAUGAUUAUUCAUUGGGCAUGGCAGCCACAAGC______CCGCACACAAG______________CACACUUGUCUGUGGCAAACAAUUUCAGCAAUUAGAAUAUCAAGCUAUGCA ..(((((((....)))))))((((.((................(((((..............................)))))........(((........)))......)).)))). (-15.59 = -14.98 + -0.62)

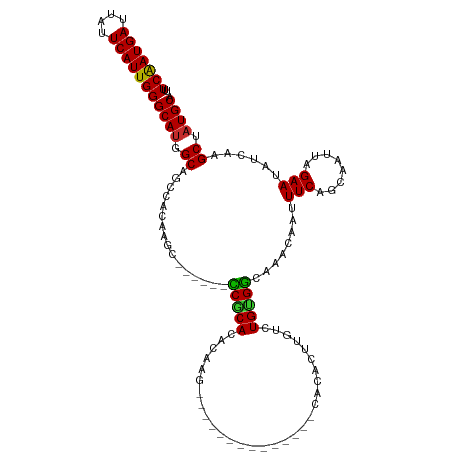
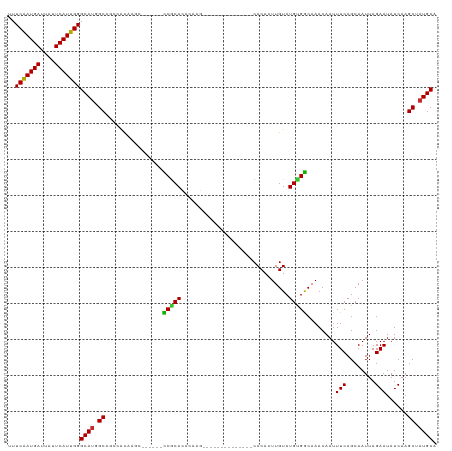
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:41 2011