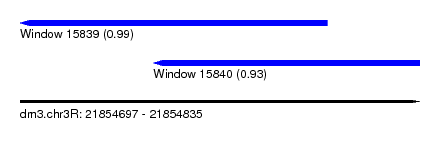
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,854,697 – 21,854,835 |
| Length | 138 |
| Max. P | 0.990961 |

| Location | 21,854,697 – 21,854,803 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Shannon entropy | 0.34076 |
| G+C content | 0.43008 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -17.36 |
| Energy contribution | -18.78 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.990961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
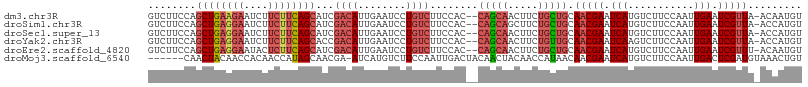
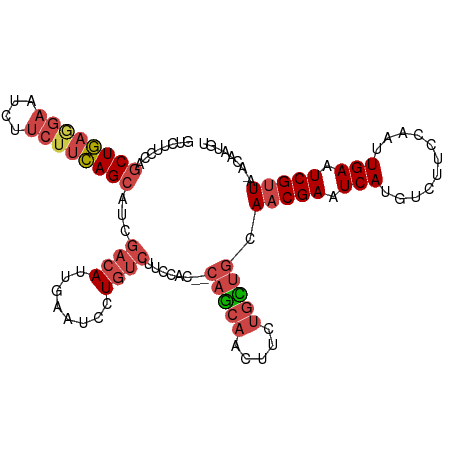
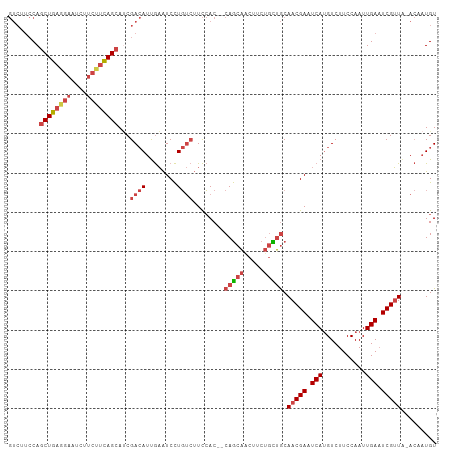
>dm3.chr3R 21854697 106 - 27905053 GUCUUCCAGCUGAAGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCAC--CAGCAACUUCUGCUGCAACGAAUCAUGUCUUCCAAUUGAAUCGUUA-ACAAUGU ........((((((((....))))))))....((((((...............--(((((.....))))).(((((.(((...........))).))))).-.)))))) ( -28.30, z-score = -3.98, R) >droSim1.chr3R 21706196 106 - 27517382 GUCUUCCAGCUGAGGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCAC--CAGCAGCUUCUGCUGCAACGAAUCAUGUCUUCCAAUUGAAUCGUUA-ACCAUGU ........((((((((....))))))))...((((........))))......--..(((((....)))))(((((.(((...........))).))))).-....... ( -29.60, z-score = -3.29, R) >droSec1.super_13 649299 106 - 2104621 GUCUUCCAGCUGAGGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCAC--CAGCAACUUCUGCUGCAACGAAUCAUGUCUUCCAAUUGAAUCGUUA-ACCAUGU ........((((((((....))))))))...((((........))))......--(((((.....))))).(((((.(((...........))).))))).-....... ( -25.60, z-score = -2.45, R) >droYak2.chr3R 4211363 106 + 28832112 GUCUUCCAGCUGAGGAAUCUUCUUCAGCACCGACAUUGAAUCCUGUCUUCCAC--CAGCAACUUCUGUUGCAACGAAUCAAGUCUUCCAAUUGAAUCGUUA-ACCAUGU ........((((((((....))))))))...((((........))))......--..(((((....)))))(((((.((((.........)))).))))).-....... ( -28.30, z-score = -3.55, R) >droEre2.scaffold_4820 4278576 106 + 10470090 GUCUUCCAGCUGAGGAAUACUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCAC--CAGCAACUUCUGCUGCAACGAAUCAUGUCUUCCAAUUGAAUCGUUU-ACAAUGU ........((((((((....))))))))....((((((...............--(((((.....))))).(((((.(((...........))).))))).-.)))))) ( -27.60, z-score = -3.12, R) >droMoj3.scaffold_6540 15810023 102 - 34148556 ------CAACUACAACCACAACCAUAGCAACGA-AUCAUGUCUUCCAAUUGACUACAACUACAACCAUAACAACGAAUCAUGUCUUCCAAUUGACUCGAUGUAAACUGU ------....................(((.(((-.....(((........)))............................(((........)))))).)))....... ( -9.00, z-score = 0.20, R) >consensus GUCUUCCAGCUGAGGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCAC__CAGCAACUUCUGCUGCAACGAAUCAUGUCUUCCAAUUGAAUCGUUA_ACAAUGU ........((((((((....))))))))...((((........))))........(((((.....))))).(((((.(((...........))).)))))......... (-17.36 = -18.78 + 1.42)
| Location | 21,854,743 – 21,854,835 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 69.37 |
| Shannon entropy | 0.42259 |
| G+C content | 0.47276 |
| Mean single sequence MFE | -14.20 |
| Consensus MFE | -9.03 |
| Energy contribution | -10.83 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21854743 92 - 27905053 ---CACCAUCACCAUCAUCAUCGACAUCGAAUCCAGUCUUCCAGCUGAAGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCC-ACCAGCAAC ---...................((((..((.((..(((.....((((((((....))))))))...)))...)).)).))))....-......... ( -17.70, z-score = -2.67, R) >droYak2.chr3R 4211409 95 + 28832112 CAUCACCACCAACAUCAUCAACGACAUCGAAUCCAGUCUUCCAGCUGAGGAAUCUUCUUCAGCACCGACAUUGAAUCCUGUCUUCC-ACCAGCAAC ......................((((..((.((..(((.....((((((((....))))))))...)))...)).)).))))....-......... ( -17.40, z-score = -2.14, R) >dp4.chr2 11602840 75 + 30794189 ---CAUUAGCGACUUCGACUACGACUACGAAUCCUGUCUUCCAACCAA-----------------CGACUACGAAUCCUGUCUUCCAACCAACAA- ---...(((((....)).))).(((...((.((..(((..........-----------------.)))...)).))..))).............- ( -7.50, z-score = -0.69, R) >consensus ___CACCACCAACAUCAUCAACGACAUCGAAUCCAGUCUUCCAGCUGA_GAAUCUUCUUCAGCA_CGACAUUGAAUCCUGUCUUCC_ACCAGCAAC ......................(((...((.((..(((.....((((((((....))))))))...)))...)).))..))).............. ( -9.03 = -10.83 + 1.81)

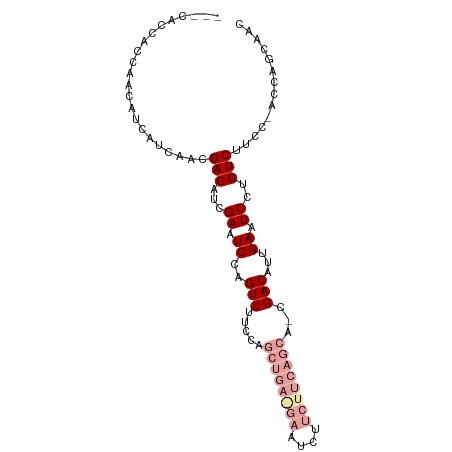
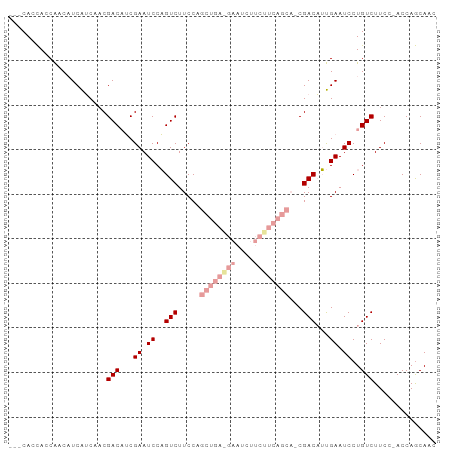
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:37 2011