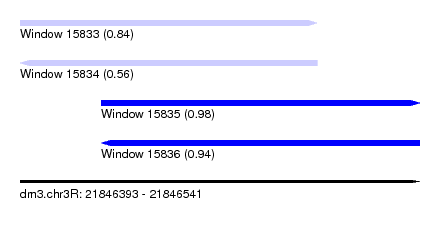
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,846,393 – 21,846,541 |
| Length | 148 |
| Max. P | 0.975039 |

| Location | 21,846,393 – 21,846,503 |
|---|---|
| Length | 110 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.81 |
| Shannon entropy | 0.10698 |
| G+C content | 0.47256 |
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -19.41 |
| Energy contribution | -19.41 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
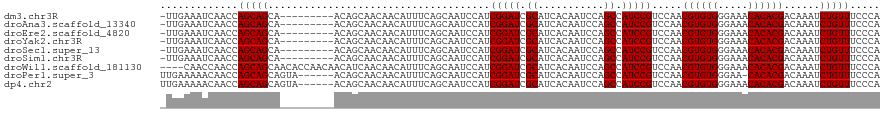
>dm3.chr3R 21846393 110 + 27905053 -UUGAAAUCAACCAGCAGCA---------ACAGCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCA -.((((((......((....---------...))......))))))(((......(((((.((...........)).))))).......)))((((((((...........)))))))). ( -23.22, z-score = -2.19, R) >droAna3.scaffold_13340 12399604 110 - 23697760 -UUGAAAUCAACCAGCAGCA---------ACAGCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCA -.((((((......((....---------...))......))))))(((......(((((.((...........)).))))).......)))((((((((...........)))))))). ( -23.22, z-score = -2.19, R) >droEre2.scaffold_4820 4269968 110 - 10470090 -UUGAAAUCAACCAGCAGCA---------ACAGCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCA -.((((((......((....---------...))......))))))(((......(((((.((...........)).))))).......)))((((((((...........)))))))). ( -23.22, z-score = -2.19, R) >droYak2.chr3R 4202553 110 - 28832112 -UUGAAAUCAACCAGCAGCA---------ACAGCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCA -.((((((......((....---------...))......))))))(((......(((((.((...........)).))))).......)))((((((((...........)))))))). ( -23.22, z-score = -2.19, R) >droSec1.super_13 640913 110 + 2104621 -UUGAAAUCAACCAGCAGCA---------ACAGCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCA -.((((((......((....---------...))......))))))(((......(((((.((...........)).))))).......)))((((((((...........)))))))). ( -23.22, z-score = -2.19, R) >droSim1.chr3R 21697528 110 + 27517382 -UUGAAAUCAACCAGCAGCA---------ACAGCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCA -.((((((......((....---------...))......))))))(((......(((((.((...........)).))))).......)))((((((((...........)))))))). ( -23.22, z-score = -2.19, R) >droWil1.scaffold_181130 15470883 116 + 16660200 ----CAACCAACCAGCAGCAACACCAACAACAUCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCA ----..........................................(((......(((((.((...........)).))))).......)))((((((((...........)))))))). ( -19.72, z-score = -2.05, R) >droPer1.super_3 5428367 113 - 7375914 UUGAAAAACAACCAGCAGCAGUA------ACAGCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAA-CACACGACAAAUCUGUUUCCCA ................(((((..------..........................(((((.((...........)).))))).....((((((....-))))))......)))))..... ( -21.30, z-score = -1.22, R) >dp4.chr2 11592498 114 - 30794189 UUGAAAAACAACCAGCAGCAGUA------ACAGCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCA .(((((........((.......------...)).......)))))(((......(((((.((...........)).))))).......)))((((((((...........)))))))). ( -20.78, z-score = -1.12, R) >consensus _UUGAAAUCAACCAGCAGCA_________ACAGCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCA .............(((((.....................................(((((.((...........)).))))).....((((((.....))))))......)))))..... (-19.41 = -19.41 + -0.00)
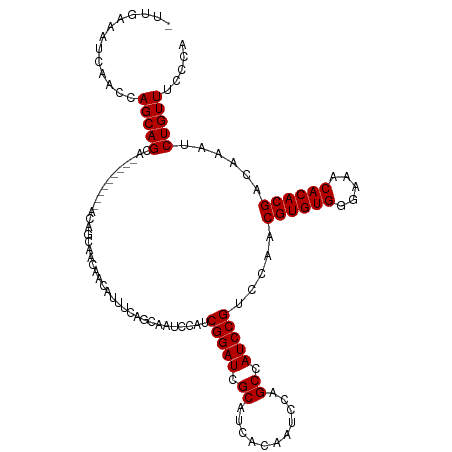
| Location | 21,846,393 – 21,846,503 |
|---|---|
| Length | 110 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.81 |
| Shannon entropy | 0.10698 |
| G+C content | 0.47256 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -29.51 |
| Energy contribution | -29.96 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.560721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21846393 110 - 27905053 UGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGCUGU---------UGCUGCUGGUUGAUUUCAA- (((((((((.((.....)).)))))))))..(((((((((.((.((......)).)).)).)))))))......((((((...((.((...---------.)).))......)))))).- ( -34.00, z-score = -1.67, R) >droAna3.scaffold_13340 12399604 110 + 23697760 UGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGCUGU---------UGCUGCUGGUUGAUUUCAA- (((((((((.((.....)).)))))))))..(((((((((.((.((......)).)).)).)))))))......((((((...((.((...---------.)).))......)))))).- ( -34.00, z-score = -1.67, R) >droEre2.scaffold_4820 4269968 110 + 10470090 UGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGCUGU---------UGCUGCUGGUUGAUUUCAA- (((((((((.((.....)).)))))))))..(((((((((.((.((......)).)).)).)))))))......((((((...((.((...---------.)).))......)))))).- ( -34.00, z-score = -1.67, R) >droYak2.chr3R 4202553 110 + 28832112 UGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGCUGU---------UGCUGCUGGUUGAUUUCAA- (((((((((.((.....)).)))))))))..(((((((((.((.((......)).)).)).)))))))......((((((...((.((...---------.)).))......)))))).- ( -34.00, z-score = -1.67, R) >droSec1.super_13 640913 110 - 2104621 UGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGCUGU---------UGCUGCUGGUUGAUUUCAA- (((((((((.((.....)).)))))))))..(((((((((.((.((......)).)).)).)))))))......((((((...((.((...---------.)).))......)))))).- ( -34.00, z-score = -1.67, R) >droSim1.chr3R 21697528 110 - 27517382 UGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGCUGU---------UGCUGCUGGUUGAUUUCAA- (((((((((.((.....)).)))))))))..(((((((((.((.((......)).)).)).)))))))......((((((...((.((...---------.)).))......)))))).- ( -34.00, z-score = -1.67, R) >droWil1.scaffold_181130 15470883 116 - 16660200 UGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGAUGUUGUUGGUGUUGCUGCUGGUUGGUUG---- (((((((((.((.....)).)))))))))...((.(.((.((..(((((((((.....)))))))((.(((.((......)).))).))))..)).)).).)).............---- ( -32.40, z-score = -1.00, R) >droPer1.super_3 5428367 113 + 7375914 UGGGAAACAGAUUUGUCGUGUG-UUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGCUGU------UACUGCUGCUGGUUGUUUUUCAA ((..((((((.(..(.((((((-....)))))).)..)(((.((((..(((.(..(((((((((...))))))).........))..).))------)...))))))).))))))..)). ( -32.50, z-score = -0.98, R) >dp4.chr2 11592498 114 + 30794189 UGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGCUGU------UACUGCUGCUGGUUGUUUUUCAA (((((((((.((.....)).))))))))).......((..((..((..(...((((((((((((...)))))))......((....)).))------))).....)..))..))..)).. ( -34.90, z-score = -1.78, R) >consensus UGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGCUGU_________UGCUGCUGGUUGAUUUCAA_ (((((((((.((.....)).)))))))))..(((((((((.((.((......)).)).)).))))))).......(((((......((.((..........)).))......)))))... (-29.51 = -29.96 + 0.45)

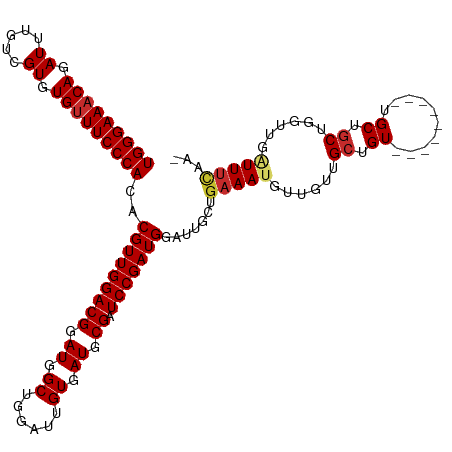
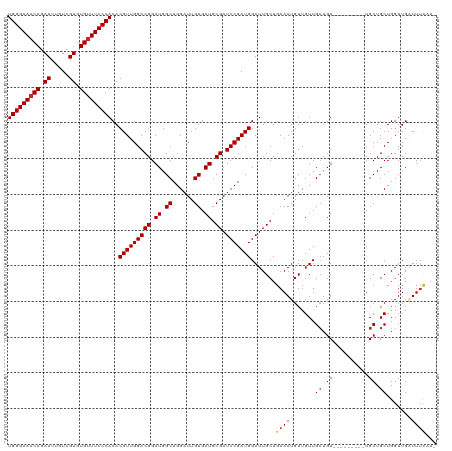
| Location | 21,846,423 – 21,846,541 |
|---|---|
| Length | 118 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.12 |
| Shannon entropy | 0.13468 |
| G+C content | 0.49607 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -25.19 |
| Energy contribution | -25.31 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21846423 118 + 27905053 AUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACACCGCA-CACACACACAAA ......((.......(((((.((...........)).)))))......((((((((((((...........)))))))))).(((((.....).))))))...)).-............ ( -27.60, z-score = -2.17, R) >droAna3.scaffold_13340 12399634 119 - 23697760 AUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACAACACAGCGCAACUAUACA ...........((..(((((.((...........)).)))))........((((((((((...........))))))))))))....(((.(((.(((........)).).))).))). ( -29.00, z-score = -2.64, R) >droEre2.scaffold_4820 4269998 118 - 10470090 AUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACACCGCA-CACACACACACA ......((.......(((((.((...........)).)))))......((((((((((((...........)))))))))).(((((.....).))))))...)).-............ ( -27.60, z-score = -2.10, R) >droSec1.super_13 640943 118 + 2104621 AUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACACCGCA-CACACACACACA ......((.......(((((.((...........)).)))))......((((((((((((...........)))))))))).(((((.....).))))))...)).-............ ( -27.60, z-score = -2.10, R) >droVir3.scaffold_13047 5427362 111 + 19223366 AUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACAACACA--ACACA------ ......((.((((...)))).))...........(((...((.....))(((((((((((...........))))))))))).............)))........--.....------ ( -25.70, z-score = -2.17, R) >droMoj3.scaffold_6540 15798646 111 + 34148556 AUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACAACACA--ACACA------ ......((.((((...)))).))...........(((...((.....))(((((((((((...........))))))))))).............)))........--.....------ ( -25.70, z-score = -2.17, R) >droGri2.scaffold_14906 10803545 111 + 14172833 AUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACAACACA--ACACA------ ......((.((((...)))).))...........(((...((.....))(((((((((((...........))))))))))).............)))........--.....------ ( -25.70, z-score = -2.17, R) >droPer1.super_3 5428401 105 - 7375914 AUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAA-CACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACCACACA------------- ...............(((((.((...........)).)))))......(((((((((-.(((.((....))))))))))))(((.((........)).)))))...------------- ( -23.60, z-score = -1.29, R) >consensus AUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACAACACA__ACACA______ ......((.((((...)))).))...........(((...((.....))(((((((((((...........))))))))))).............)))..................... (-25.19 = -25.31 + 0.12)

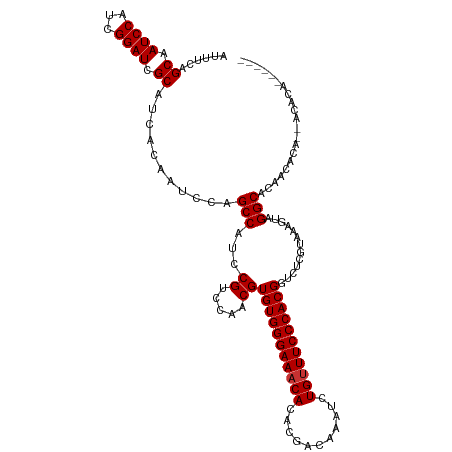
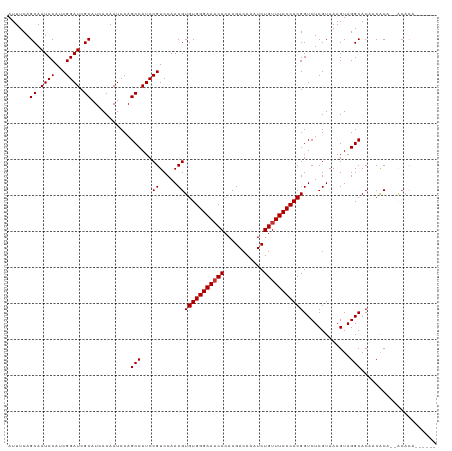
| Location | 21,846,423 – 21,846,541 |
|---|---|
| Length | 118 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.12 |
| Shannon entropy | 0.13468 |
| G+C content | 0.49607 |
| Mean single sequence MFE | -38.39 |
| Consensus MFE | -33.39 |
| Energy contribution | -33.51 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21846423 118 - 27905053 UUUGUGUGUGUG-UGCGGUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAU ((((((...(((-.((.....)).)))..))))))...((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))............ ( -39.30, z-score = -1.87, R) >droAna3.scaffold_13340 12399634 119 + 23697760 UGUAUAGUUGCGCUGUGUUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAU .(((.(((.((((......))))..))).)))......((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))............ ( -42.90, z-score = -3.30, R) >droEre2.scaffold_4820 4269998 118 + 10470090 UGUGUGUGUGUG-UGCGGUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAU .((.((((...(-((.((....)))))..))))..)).((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))............ ( -38.20, z-score = -1.39, R) >droSec1.super_13 640943 118 - 2104621 UGUGUGUGUGUG-UGCGGUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAU .((.((((...(-((.((....)))))..))))..)).((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))............ ( -38.20, z-score = -1.39, R) >droVir3.scaffold_13047 5427362 111 - 19223366 ------UGUGU--UGUGUUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAU ------.((.(--((((..((....))..))))).)).((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))............ ( -39.10, z-score = -3.01, R) >droMoj3.scaffold_6540 15798646 111 - 34148556 ------UGUGU--UGUGUUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAU ------.((.(--((((..((....))..))))).)).((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))............ ( -39.10, z-score = -3.01, R) >droGri2.scaffold_14906 10803545 111 - 14172833 ------UGUGU--UGUGUUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAU ------.((.(--((((..((....))..))))).)).((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))............ ( -39.10, z-score = -3.01, R) >droPer1.super_3 5428401 105 + 7375914 -------------UGUGUGGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUG-UUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAU -------------...(..((.................((((((((((((....)).))).-))))))).(((((((((.((.((......)).)).)).))))))).))..)...... ( -31.20, z-score = -0.74, R) >consensus ______UGUGU__UGUGUUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAU .....................((...............((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))....))...... (-33.39 = -33.51 + 0.12)

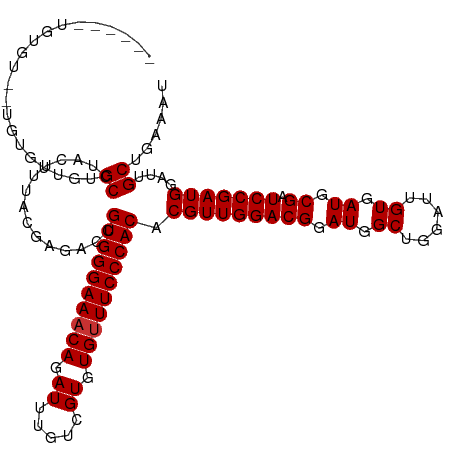
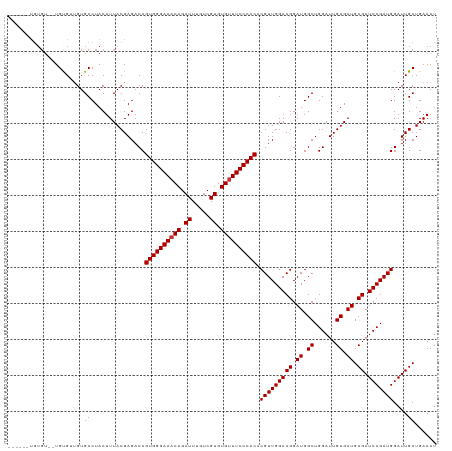
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:34 2011