
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,843,133 – 21,843,290 |
| Length | 157 |
| Max. P | 0.558506 |

| Location | 21,843,133 – 21,843,235 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.73 |
| Shannon entropy | 0.35648 |
| G+C content | 0.55333 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
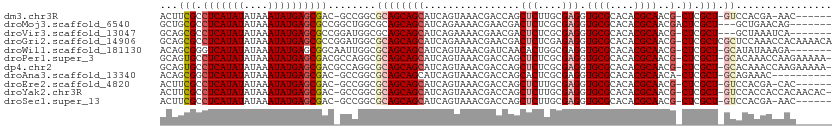
>dm3.chr3R 21843133 102 - 27905053 ACUUCGCCUCAUAUAUAAAUAUGAGCGAC-GCCGGCGCAGCAGCAUCAGUAAACGACCAGCUCUUGCGAGGUGCGCACACGCAACG-CUCGCU-GUCCACGA-AAC------ ...((((.((((((....))))))))))(-(..((.(((((.(((..(((.........)))..)))((((((((....))).)).-))))))-)))).)).-...------ ( -32.60, z-score = -1.29, R) >droMoj3.scaffold_6540 15794580 102 - 34148556 GCUGCGCCUCAUAUAUAAAUAUGAGCGCCGGCUGGCGCAGCAGCAUCAGAAAACGAACGACUCUCGCGAGGUGCGCACACGCAACGACUCGCU---GCUGAACAG------- (((((...((((((....))))))(((((....))))).))))).((((.....((.......))(((((.((((....))))....))))).---.))))....------- ( -39.50, z-score = -2.46, R) >droVir3.scaffold_13047 5423467 101 - 19223366 GCAGCGCCUCAUAUAUAAAUAUGAGCGCCGGAUGGCGCAGCAGCAUCAGAAAACGAACGACUCUCGCGAGGUGCGCACACGCAACG-CUCGCU---GCUAAAUCA------- (((((((.((((((....))))))(((((....))))).))(((................(((....))).((((....))))..)-)).)))---)).......------- ( -33.40, z-score = -0.81, R) >droGri2.scaffold_14906 10799566 111 - 14172833 GCAGCGCCUCAUAUAUAAAUAUGAGCGCCGGAUGGCGCAGCAGCAUCAGAAAACGAACGACUCUCGAGAGGUGCGCACACGCAACG-CUCGCUCGCUCCAAACCACAAAACA ...((((.((((((....)))))))))).((.(((.((((((((................(((....))).((((....))))..)-)).))).)).)))..))........ ( -33.60, z-score = -1.56, R) >droWil1.scaffold_181130 15467197 103 - 16660200 ACAGCGGGUCAUAUAUAAAUAUGAGCGGCAAUUGGCGCAGCAGCAUCAGUAAACGAUCAACACUGGCGAGGUGCGCACACGCAACG-CUCGCU-GCAUAUAAAGA------- .(((((((((((((....))))))(((.......(((((.(.((..((((...........))))))..).)))))...)))....-))))))-)..........------- ( -31.50, z-score = -1.31, R) >droPer1.super_3 5424891 109 + 7375914 GCAGUGCCUCAUAUAUAAAUAUGAGCGACGCCAGGCGCAGCAGCAUCAGUAAACGACCAGCUCUCGCGAGGUGCGCACACGCAACG-CUCGCU-GCACAAACCAAGAAAAA- ((.(((((((((((....))))))).).)))...))((((((((...........(((.((....))..)))(((....)))...)-)).)))-))...............- ( -33.00, z-score = -1.10, R) >dp4.chr2 11589147 109 + 30794189 GCAGUGCCUCAUAUAUAAAUAUGAGCGACGCCAGGCGCAGCAGCAUCAGUAAACGACCAGCUCUCGCGAGGUGCGCACACGCAACG-CUCGCU-GCACAAACCAAGAAAAA- ((.(((((((((((....))))))).).)))...))((((((((...........(((.((....))..)))(((....)))...)-)).)))-))...............- ( -33.00, z-score = -1.10, R) >droAna3.scaffold_13340 12395841 99 + 23697760 ACAGCGGCUCAUAUAUAAAUAUGAGCGAC-GCCGGCGCAGCAGCAUCAGUAAACGACCAGCACUCGCGAGGUGCGCACACGCAACA-CUCGCU-GCAGAAAC---------- .(.(((((((((((....))))))))..)-)).)..(((((..................((....))(((.((((....))))...-))))))-))......---------- ( -33.10, z-score = -1.85, R) >droEre2.scaffold_4820 4266706 102 + 10470090 ACUUCGCCUCAUAUAUAAAUAUGAGCGAC-GCCGGCGCAGCAGCAUCAGUAAACGACCAGCUCUUGCGAGGUGCGCACACGCAACG-CUCGCU-GUCCACGA-CAC------ ...((((.((((((....))))))))))(-(..((.(((((.(((..(((.........)))..)))((((((((....))).)).-))))))-)))).)).-...------ ( -32.60, z-score = -1.28, R) >droYak2.chr3R 4199281 108 + 28832112 ACUUCGCCUCAUAUAUAAAUAUGAGCGAC-GCCGGCGCAGCAGCAUCAGUAAACGACCAGCUCUUGCGAGGUGCGCACACGCAACG-CUCGCU-GUCCACCACCACAACAC- ...((((.((((((....)))))))))).-...((.(((((.(((..(((.........)))..)))((((((((....))).)).-))))))-)))).............- ( -32.30, z-score = -1.83, R) >droSec1.super_13 637636 102 - 2104621 ACUUCGCCUCAUAUAUAAAUAUGAGCGAC-GCCGGCGCAGCAGCAUCAGUAAACGACCAGCUCUUGCGAGGUGCGCACACGCAACG-CUCGCU-GUCCACGA-AAC------ ...((((.((((((....))))))))))(-(..((.(((((.(((..(((.........)))..)))((((((((....))).)).-))))))-)))).)).-...------ ( -32.60, z-score = -1.29, R) >consensus ACAGCGCCUCAUAUAUAAAUAUGAGCGAC_GCCGGCGCAGCAGCAUCAGUAAACGACCAGCUCUCGCGAGGUGCGCACACGCAACG_CUCGCU_GCACAAAACAAC______ ...(((.(((((((....))))))))))......((......)).....................(((((.((((....))))....))))).................... (-21.70 = -21.90 + 0.20)
| Location | 21,843,196 – 21,843,290 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.11 |
| Shannon entropy | 0.27216 |
| G+C content | 0.56819 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -26.46 |
| Energy contribution | -26.66 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.558506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
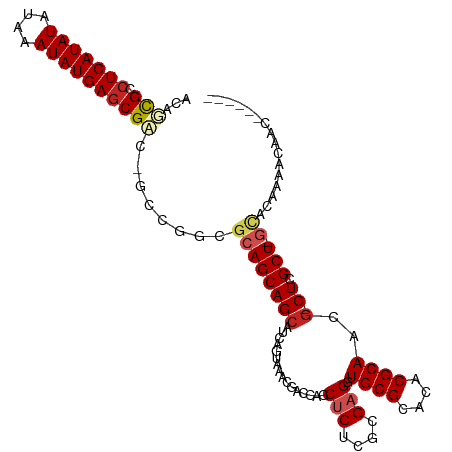
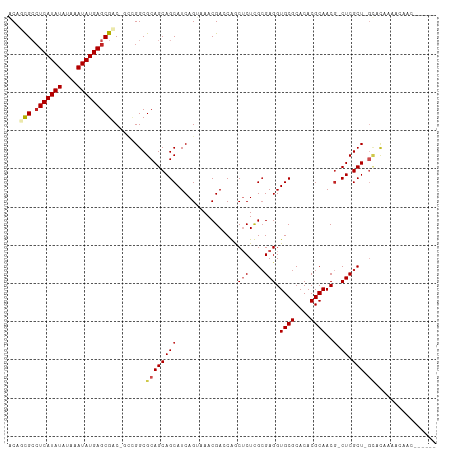
>dm3.chr3R 21843196 94 - 27905053 -------CGUGAGAUCAGCCGGCGUACGAUUUCACUCUUGACUGGCCAUCGGCA------------------GCGCAGCAACUUCGCCUCAUAUAUAAAUAUGAGCGACGCC-GGCGCAG -------(....)....((((((((............(((.(((((........------------------)).))))))...(((.((((((....))))))))))))))-))).... ( -32.40, z-score = -1.56, R) >droMoj3.scaffold_6540 15794642 95 - 34148556 -------CGUAAGAUCAGCCGGCAUACGAUUUCACUCUUGACUGGCCAUUGGCA------------------GCGCUGCGGCUGCGCCUCAUAUAUAAAUAUGAGCGCCGGCUGGCGCAG -------.....(.(((((((((............................(((------------------((......)))))((.((((((....)))))))))))))))))).... ( -35.00, z-score = -0.94, R) >droVir3.scaffold_13047 5423528 95 - 19223366 -------CGUGAGAUCAGCCGGCAUACGAUUUCACUCUUGACUGGCCAUUGGCA------------------GCGCUGCGGCAGCGCCUCAUAUAUAAAUAUGAGCGCCGGAUGGCGCAG -------.((((((((...........))))))))......((((((((((((.------------------((((((...))))))(((((((....))))))).))).)))))).))) ( -40.00, z-score = -2.61, R) >droGri2.scaffold_14906 10799637 95 - 14172833 -------CGUGAGAUCAGCCCGCAGACGAUUUCACUCUUGACUGGCCAUUGGCA------------------GCGCUGCGGCAGCGCCUCAUAUAUAAAUAUGAGCGCCGGAUGGCGCAG -------.((((((((...........))))))))......((((((((((((.------------------((((((...))))))(((((((....))))))).))).)))))).))) ( -40.00, z-score = -2.64, R) >droWil1.scaffold_181130 15467260 108 - 16660200 -------CGUGAGAUCAGCAG-----CGAUUUCACUCUUGACUGGCCAUUGGCAGCAGAGCAGAGCAGCGCUGCGCAGCAACAGCGGGUCAUAUAUAAAUAUGAGCGGCAAUUGGCGCAG -------.((((((((.....-----.))))))))......(((((((...((.((((.((......)).)))))).......((.(.((((((....)))))).).))...)))).))) ( -39.90, z-score = -1.69, R) >droPer1.super_3 5424960 101 + 7375914 -GCUGCUCGUGAGAUCAGCCGGCAUACGAUUUCACUCUUGACUGGCCAUCGGCU------------------GCGCAGCGGCAGUGCCUCAUAUAUAAAUAUGAGCGACGCCAGGCGCAG -((((.((....)).)))).(((...(((........)))....))).....((------------------((((...(((..(((.((((((....)))))))))..)))..)))))) ( -37.80, z-score = -1.14, R) >dp4.chr2 11589216 101 + 30794189 -GCUGCUCGUGAGAUCAGCCGGCAUACGAUUUCACUCUUGACUGGCCAUCGGCU------------------GCGCAGCGGCAGUGCCUCAUAUAUAAAUAUGAGCGACGCCAGGCGCAG -((((.((....)).)))).(((...(((........)))....))).....((------------------((((...(((..(((.((((((....)))))))))..)))..)))))) ( -37.80, z-score = -1.14, R) >droAna3.scaffold_13340 12395901 101 + 23697760 UGGUGCUCGUGAGAUCAGCCGGCAUACGAUUUCACUCUUGACUGGCCAGCGGCA------------------GCGCAGCGACAGCGGCUCAUAUAUAAAUAUGAGCGACGCC-GGCGCAG ...(((((....))...((((((.........(.((.(((.(((((........------------------)).)))))).)).)((((((((....))))))))...)))-)))))). ( -37.30, z-score = -0.88, R) >droEre2.scaffold_4820 4266769 94 + 10470090 -------CGUGAGAUCAGCCGGCGUACGAUUUCACUCUUGACUGGCCAUCGGCA------------------GCGCAGCAACUUCGCCUCAUAUAUAAAUAUGAGCGACGCC-GGCGCAG -------(....)....((((((((............(((.(((((........------------------)).))))))...(((.((((((....))))))))))))))-))).... ( -32.40, z-score = -1.56, R) >droYak2.chr3R 4199350 94 + 28832112 -------CGUGAGAUCAGCCGGCGUACGAUUUCACUCUUGACUGGCCAUCGGCA------------------GCGCAGCAACUUCGCCUCAUAUAUAAAUAUGAGCGACGCC-GGCGCAG -------(....)....((((((((............(((.(((((........------------------)).))))))...(((.((((((....))))))))))))))-))).... ( -32.40, z-score = -1.56, R) >droSec1.super_13 637699 94 - 2104621 -------CGUGAGAUCAGCCGGCGUACGAUUUCACUCUUGACUGGCCAUCGGCA------------------GCGCAGCAACUUCGCCUCAUAUAUAAAUAUGAGCGACGCC-GGCGCAG -------(....)....((((((((............(((.(((((........------------------)).))))))...(((.((((((....))))))))))))))-))).... ( -32.40, z-score = -1.56, R) >consensus _______CGUGAGAUCAGCCGGCAUACGAUUUCACUCUUGACUGGCCAUCGGCA__________________GCGCAGCGACAGCGCCUCAUAUAUAAAUAUGAGCGACGCC_GGCGCAG ........((((((((...........)))))))).........(((...)))...................((((.......(((.(((((((....))))))))))......)))).. (-26.46 = -26.66 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:29 2011