
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,838,452 – 21,838,559 |
| Length | 107 |
| Max. P | 0.955452 |

| Location | 21,838,452 – 21,838,559 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.89 |
| Shannon entropy | 0.50852 |
| G+C content | 0.43253 |
| Mean single sequence MFE | -32.01 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.14 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21838452 107 - 27905053 CUUCGCUUGCUCCU------CCUGUGCUGUCGACAUCAAUUUGCAUUUUUAGUGUAGGAGAAAUGCGUUUGAGAUUGAUAGAGAAGGACGGACAGAGUGU------GUGAAAGAGUGGG ...(((((..((((------(((...(((((((..((((..(((((((((........))))))))).))))..)))))))...)))).)))..))))).------............. ( -34.60, z-score = -2.53, R) >droPer1.super_3 5419945 104 + 7375914 UUCUUUCUACUCCUU-----CUCAUGCUGUCGACAUCAAUUUGCAUUUUUAGUGUAGGAGAAAUGCGUUUGUGAUUGAUAGAAAGAGACAGGGUGUGAGAGAGAAUAAU---------- (((((((((((((((-----(((...(((((((((.(((..(((((((((........))))))))).))))).)))))))...)))).)))).)).))))))))....---------- ( -35.40, z-score = -3.92, R) >dp4.chr2 11584237 104 + 30794189 UUCUUUCUACUCCUU-----CUCAUGCUGUCGACAUCAAUUUGCAUUUUUAGUGUAGGAGAAAUGCGUUUGUGAUUGAUAGAAAGAGACAGGGUGUGAGAGAGAAUAAU---------- (((((((((((((((-----(((...(((((((((.(((..(((((((((........))))))))).))))).)))))))...)))).)))).)).))))))))....---------- ( -35.40, z-score = -3.92, R) >droAna3.scaffold_13340 12390904 119 + 23697760 CUUCUUGAGACCCUCUAUAGCCCCACCUGUCGACAUCAAUUUGCAUAUUUAGUGUAGAAGAAAUGCGUUUGUGAUUGAUAGUGAGAUGGAGGCGGGGAGCGGGCCAGUGAAAGAGUGGG ..........((((((...(((((.((((((..((((..(((((((.....)))))))......((....))............))))..))))))..).)))).......)))).)). ( -32.90, z-score = -0.48, R) >droEre2.scaffold_4820 4262004 107 + 10470090 CUUCGUUUGCUCCU------CCCCUGCUGUCGACAUCAAUUUGCAUUUUUAGUGUAGGAAAAAUGCGUUUGAGAUUGAUAGAGAAGGACGGACAGAGUGU------GUGAAAGAGUCGG .((((..(((((.(------(((((.(((((((..((((..(((((((((........))))))))).))))..)))))))...)))..)))..))))).------.))))........ ( -32.40, z-score = -2.20, R) >droYak2.chr3R 4191225 107 + 28832112 CUUCGUUUGCUCCU------CCCCGGCUGUCGACAUCAAUUUGUAUUUUUAGUGUAGGAGAAAUGCGUUUGAGAUUGAUAGAGAAGGACGGACAGAGUGU------GUGAAAGAGUGGG .((((..(((((.(------((((..(((((((..((((..(((((((((........))))))))).))))..)))))))....))..)))..))))).------.))))........ ( -28.80, z-score = -1.13, R) >droSec1.super_13 632787 107 - 2104621 CUUCGCUUGCUCCU------CCCGUGCUGUCGACAUCAAUUUGCAUUUUUAGUGUAGGAGAAAUGCGUUUGAGAUUGAUAGAGAAGGACGGACAGAGUGU------GUGAAAGAGUGGG .(((((.(((((..------.((((.(((((((..((((..(((((((((........))))))))).))))..)))))))......))))...))))).------)))))........ ( -32.50, z-score = -1.83, R) >droSim1.chr3R 21690161 107 - 27517382 CUUCGCUUGCUCCU------CCCGUGCUGUCGACAUCAAUUUGCAUUUUUAGUGUAGGAGAAAUGCGUUUGAGAUUGAUAGAGAAGGACGGACAGAGUGU------GUGAAAGAGUGGG .(((((.(((((..------.((((.(((((((..((((..(((((((((........))))))))).))))..)))))))......))))...))))).------)))))........ ( -32.50, z-score = -1.83, R) >droVir3.scaffold_13047 5418029 87 - 19223366 ----UUCUACUUCUU-----UUU---UUUUUGACAUCAAUUUGCAUUUUUAGUGUAGGAGAAAUGCGUUAGUGAUUGAUAGAGCGAAAGAGCGAGAGGG-------------------- ----.....((((((-----.((---((((((..((((((((((((((((........))))))))).....)))))))....)))))))).)))))).-------------------- ( -23.60, z-score = -2.54, R) >consensus CUUCGUUUGCUCCU______CCCCUGCUGUCGACAUCAAUUUGCAUUUUUAGUGUAGGAGAAAUGCGUUUGAGAUUGAUAGAGAAGGACGGACAGAGUGU______GUGAAAGAGUGGG ....................((....(((((((...(((..(((((((((........))))))))).)))...)))))))....))................................ (-14.52 = -14.14 + -0.38)

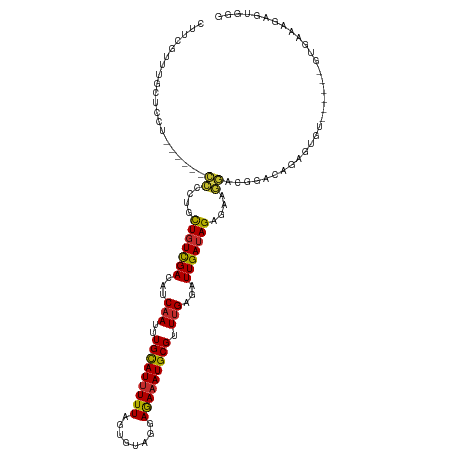
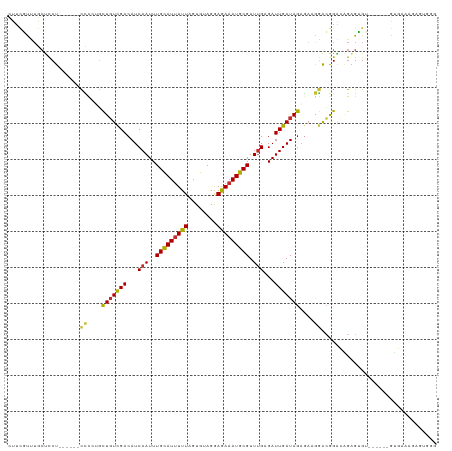
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:28 2011