
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,714,167 – 21,714,272 |
| Length | 105 |
| Max. P | 0.767786 |

| Location | 21,714,167 – 21,714,272 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 59.44 |
| Shannon entropy | 0.78316 |
| G+C content | 0.36601 |
| Mean single sequence MFE | -20.89 |
| Consensus MFE | -4.95 |
| Energy contribution | -4.45 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.767786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
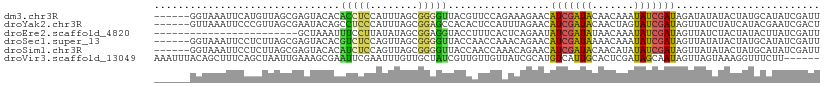
>dm3.chr3R 21714167 105 + 27905053 ------GGUAAAUUCAUGUUAGCGAGUACACACCUCCAUUUAGCGGGGUUACGUUCCAGAAAGAACAUCGAUACAACAAAUAUCGAUAGAUAUAUACUAUGCAUAUCGAUU ------..........(((...((((((...((((((.....).))))))))((((......)))).)))..)))......(((((((..((((...))))..))))))). ( -19.70, z-score = -1.03, R) >droYak2.chr3R 4064907 105 - 28832112 ------GUUAAAUUCCCGUUAGCGAAUACAGCCUCCCAUUUAGCGGAGCCACACUCCAUUUAGAACAUCGAUACAACUAGUAUCGAUAGUUAUCUAUCAUACGAAUCGACU ------(((((((........((.......)).....)))))))((((.....))))...((((((((((((((.....)))))))).))..))))............... ( -20.62, z-score = -2.48, R) >droEre2.scaffold_4820 4135112 87 - 10470090 ------------------------GCUAAAUUUCCUUAUAUAGCGGAGGUACCUUUCACUCAGAAUAUCGAUAUAACAAAUAUCGAUAGUUAUCUACUAUACUUAUCGAUU ------------------------..............(((((..((...((..(((.....)))(((((((((.....)))))))))))..))..))))).......... ( -14.90, z-score = -1.50, R) >droSec1.super_13 497497 105 + 2104621 ------GGUAAAUUCCUCUUAGCGAGUACACGUCUCCAGUUAGCGGGGUUACCAACCAAACAGAACAUCGAUAAAACAAAUAUCGAUAGUUAUAUACUAUGCAUAUCGAUU ------(((((...((((((((((((.......)))..))))).)))))))))..........((((((((((.......))))))).))).................... ( -20.30, z-score = -1.41, R) >droSim1.chr3R 21558914 105 + 27517382 ------GGUAAAUUCCUCUUAGCGAGUACACAUCUCCAGUUAGCGGGGUUACCAACCAAACAGAACAUCGAUACAACAUAUAUCGAUAGUUAUAUACUAUGCAUAUCGAUU ------(((((...((((((((((((.......)))..))))).)))))))))..........((((((((((.......))))))).))).................... ( -20.10, z-score = -1.61, R) >droVir3.scaffold_13049 17206875 105 - 25233164 AAAUUUACAGCUUUCAGCUAAUUGAAAGCGAAUUCGAAUUUGUUGCUAUCGUUGUUGUUAUCGCAUGUCAUUGCACUCGAUAGCAAUAGUUAGUAAAGGUUUCUU------ .........((((((((....))))))))......(((....((((((...((((((((((((..(((....)))..)))))))))))).))))))....)))..------ ( -29.70, z-score = -3.24, R) >consensus ______GGUAAAUUCCUCUUAGCGAGUACACAUCCCCAUUUAGCGGGGUUACCAUCCAAACAGAACAUCGAUACAACAAAUAUCGAUAGUUAUAUACUAUGCAUAUCGAUU ...............................(((((........))))).................(((((((.......)))))))........................ ( -4.95 = -4.45 + -0.50)

| Location | 21,714,167 – 21,714,272 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 59.44 |
| Shannon entropy | 0.78316 |
| G+C content | 0.36601 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -6.32 |
| Energy contribution | -6.27 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.563525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
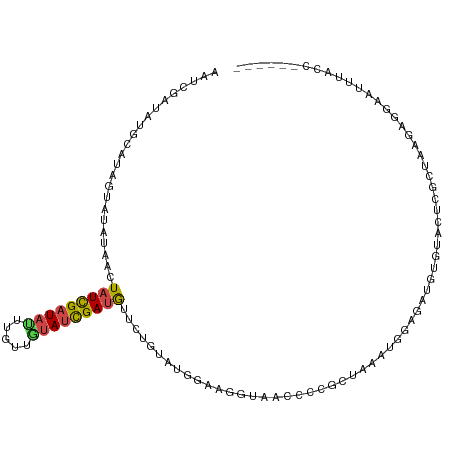
>dm3.chr3R 21714167 105 - 27905053 AAUCGAUAUGCAUAGUAUAUAUCUAUCGAUAUUUGUUGUAUCGAUGUUCUUUCUGGAACGUAACCCCGCUAAAUGGAGGUGUGUACUCGCUAACAUGAAUUUACC------ ..(((....((..(((((((((((((((((((.....))))))))(((((....)))))......(((.....)))))))))))))).)).....))).......------ ( -25.70, z-score = -1.90, R) >droYak2.chr3R 4064907 105 + 28832112 AGUCGAUUCGUAUGAUAGAUAACUAUCGAUACUAGUUGUAUCGAUGUUCUAAAUGGAGUGUGGCUCCGCUAAAUGGGAGGCUGUAUUCGCUAACGGGAAUUUAAC------ ......(((((....((((....(((((((((.....))))))))).))))....(((((..(((((........))).))..)))))....)))))........------ ( -29.40, z-score = -1.93, R) >droEre2.scaffold_4820 4135112 87 + 10470090 AAUCGAUAAGUAUAGUAGAUAACUAUCGAUAUUUGUUAUAUCGAUAUUCUGAGUGAAAGGUACCUCCGCUAUAUAAGGAAAUUUAGC------------------------ .........((((((((((....(((((((((.....))))))))).)))........((.....))))))))).............------------------------ ( -16.40, z-score = -0.64, R) >droSec1.super_13 497497 105 - 2104621 AAUCGAUAUGCAUAGUAUAUAACUAUCGAUAUUUGUUUUAUCGAUGUUCUGUUUGGUUGGUAACCCCGCUAACUGGAGACGUGUACUCGCUAAGAGGAAUUUACC------ ..((.....((..(((((((...((((((((.......)))))))).....((..((((((......))))))..))...))))))).))...))..........------ ( -23.20, z-score = -0.69, R) >droSim1.chr3R 21558914 105 - 27517382 AAUCGAUAUGCAUAGUAUAUAACUAUCGAUAUAUGUUGUAUCGAUGUUCUGUUUGGUUGGUAACCCCGCUAACUGGAGAUGUGUACUCGCUAAGAGGAAUUUACC------ ..((.....((..((((((((..(((((((((.....)))))))))...(.((..((((((......))))))..)).))))))))).))...))..........------ ( -25.50, z-score = -1.22, R) >droVir3.scaffold_13049 17206875 105 + 25233164 ------AAGAAACCUUUACUAACUAUUGCUAUCGAGUGCAAUGACAUGCGAUAACAACAACGAUAGCAACAAAUUCGAAUUCGCUUUCAAUUAGCUGAAAGCUGUAAAUUU ------........(((((......(((((((((...(((......)))(....).....))))))))).............(((((((......))))))).)))))... ( -23.10, z-score = -2.76, R) >consensus AAUCGAUAUGCAUAGUAUAUAACUAUCGAUAUUUGUUGUAUCGAUGUUCUGUAUGGAAGGUAACCCCGCUAAAUGGAGAUGUGUACUCGCUAAGAGGAAUUUACC______ .......................(((((((((.....)))))))))................................................................. ( -6.32 = -6.27 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:20 2011