| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,708,648 – 21,708,739 |
| Length | 91 |
| Max. P | 0.927486 |

| Location | 21,708,648 – 21,708,739 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 72.65 |
| Shannon entropy | 0.45720 |
| G+C content | 0.28326 |
| Mean single sequence MFE | -18.89 |
| Consensus MFE | -11.11 |
| Energy contribution | -14.30 |
| Covariance contribution | 3.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
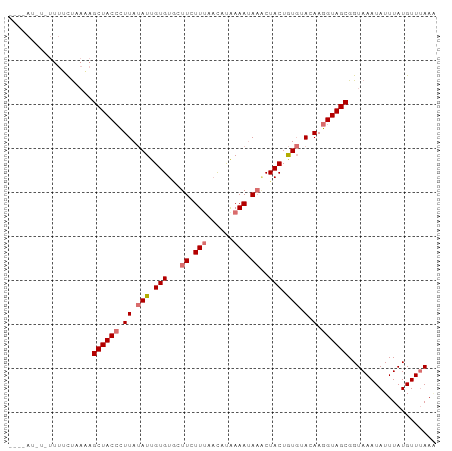
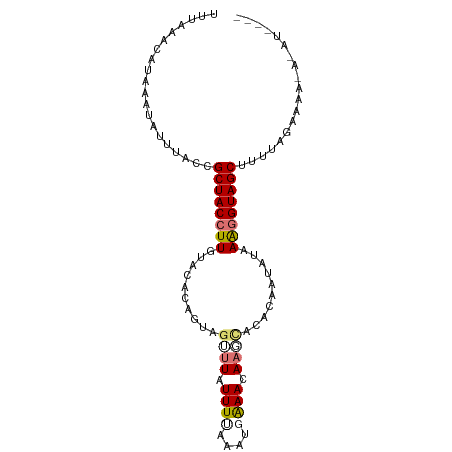
>dm3.chr3R 21708648 91 + 27905053 ----ACAGCUUUUCUAAAAGCUACCCUUAUAUUGUGU--UUCUUUCUCACGAAAUAAACUACUGUGUAGAAGGUAGCGGUAAAUAUUUAUGUUUAAA ----...............((((((.((((((.((((--((.((((....)))).)))).)).))))))..))))))..((((((....)))))).. ( -19.20, z-score = -2.03, R) >droSec1.super_13 491901 96 + 2104621 ACAUAUGUUUUUGCUAACAGCUACCCUUAUAUUGUGUAUUUCUUUCACAUAAAAAAAACUACUGUGUAAAAGGUAGCGGUAAAUAUUUAUGUUAAA- (((((.((.((((((....((((((.((((((.((((.(((.((......)).))).)).)).))))))..)))))))))))).)).)))))....- ( -19.30, z-score = -1.85, R) >droEre2.scaffold_4820 4129514 93 - 10470090 --CUAUUUAUUUUUCCAAAGCUACAUUUAUAUUGUGUGCUUGUUCAAUUUAAAAUA--CUACUGUGCACAAGGUAGCGGUAAAUAUUUAUGUUUAAA --.((..(((((..((...(((((.......(((((..(..((.............--..)).)..))))).))))))).)))))..))........ ( -15.46, z-score = -0.47, R) >droYak2.chr3R 4059291 84 - 28832112 -------------CUUUAGGCUACCUUUACAUUGUGUGCUUGUUUAAUUUAAAAUAAGCUACUAUUAACAAGGUAGCGGUAAAUAUUUAUGUUUAAG -------------......((((((((......(((.((((((((......))))))))))).......))))))))..((((((....)))))).. ( -21.62, z-score = -2.82, R) >consensus ____AU_U_UUUUCUAAAAGCUACCCUUAUAUUGUGUGCUUCUUUAACAUAAAAUAAACUACUGUGUACAAGGUAGCGGUAAAUAUUUAUGUUUAAA ...................(((((((((((((.(((.((((((((......))))))))))).))))).)))))))).................... (-11.11 = -14.30 + 3.19)


| Location | 21,708,648 – 21,708,739 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 72.65 |
| Shannon entropy | 0.45720 |
| G+C content | 0.28326 |
| Mean single sequence MFE | -16.25 |
| Consensus MFE | -9.98 |
| Energy contribution | -10.97 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
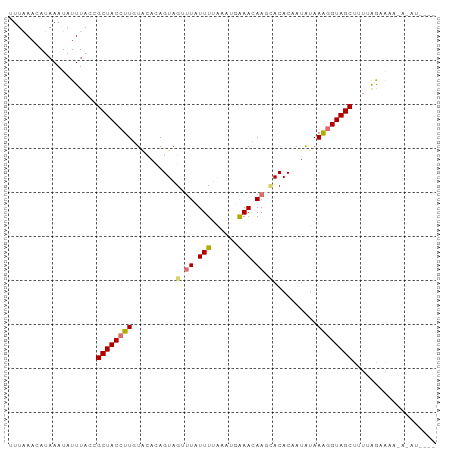
>dm3.chr3R 21708648 91 - 27905053 UUUAAACAUAAAUAUUUACCGCUACCUUCUACACAGUAGUUUAUUUCGUGAGAAAGAA--ACACAAUAUAAGGGUAGCUUUUAGAAAAGCUGU---- ....................((((((((.......((.((((.((((....)))).))--))))......))))))))...............---- ( -19.12, z-score = -2.41, R) >droSec1.super_13 491901 96 - 2104621 -UUUAACAUAAAUAUUUACCGCUACCUUUUACACAGUAGUUUUUUUUAUGUGAAAGAAAUACACAAUAUAAGGGUAGCUGUUAGCAAAAACAUAUGU -....(((((..........(((((((((......(((.(((((((.....))))))).))).......)))))))))((((......))))))))) ( -20.02, z-score = -2.49, R) >droEre2.scaffold_4820 4129514 93 + 10470090 UUUAAACAUAAAUAUUUACCGCUACCUUGUGCACAGUAG--UAUUUUAAAUUGAACAAGCACACAAUAUAAAUGUAGCUUUGGAAAAAUAAAUAG-- ............((((((..(((((..(((((....(((--(.......)))).....)))))..........)))))(((....))))))))).-- ( -10.10, z-score = 0.54, R) >droYak2.chr3R 4059291 84 + 28832112 CUUAAACAUAAAUAUUUACCGCUACCUUGUUAAUAGUAGCUUAUUUUAAAUUAAACAAGCACACAAUGUAAAGGUAGCCUAAAG------------- ....................((((((((.......((.((((.(((......))).))))))........))))))))......------------- ( -15.76, z-score = -2.12, R) >consensus UUUAAACAUAAAUAUUUACCGCUACCUUGUACACAGUAGUUUAUUUUAAAUGAAACAAGCACACAAUAUAAAGGUAGCUUUUAGAAAA_A_AU____ ....................((((((((..........((((.((((....)))).))))..........))))))))................... ( -9.98 = -10.97 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:18 2011