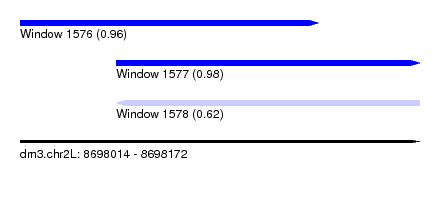
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,698,014 – 8,698,172 |
| Length | 158 |
| Max. P | 0.979304 |

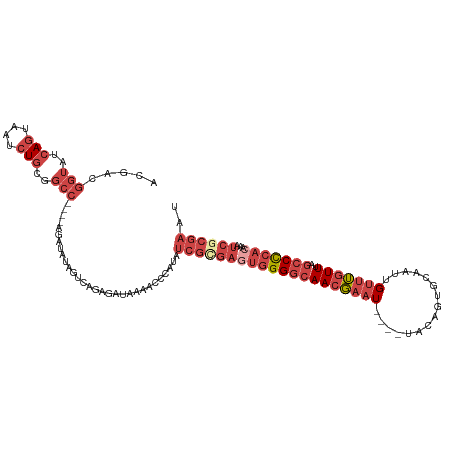
| Location | 8,698,014 – 8,698,132 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.53 |
| Shannon entropy | 0.41881 |
| G+C content | 0.44473 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -17.25 |
| Energy contribution | -19.83 |
| Covariance contribution | 2.59 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8698014 118 + 23011544 ACGACGGUAUCAGUAAUCUGCGGCCAGAUAGAUAUCAUCAGAGAUAAAAUCCAUAUCGCGAGUGGGGCAACGAAUGAAUUACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAU ..((.((((((....(((((....))))).)))))).))................(((((((((((((((((((..(............)..))))))..)))))))...)))))).. ( -36.80, z-score = -3.38, R) >droSim1.chr2L 8483259 114 + 22036055 ACGACGGUAUCAGUAAUCUGCGGCCAGAUAGAUAUUGUCAGAGAUAAAACCCAUAUCGCGAGUGGGGCAACGAAU----UACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAU ..(((((((((....(((((....))))).)))))))))................((((((((((((((((((((----............)))))))..)))))))...)))))).. ( -39.90, z-score = -4.54, R) >droSec1.super_3 4179766 114 + 7220098 ACGACGGUAUCAGUAAUCUGCGGCCAUAUAGAUAUUGUCAGAGAUAAAAUCCAUAUCGCGAGUGGGGCAACGAAU----UACAGUGCAAUUGUUUGUUAAGCCCCACAAAUCGCGAAU ..(((((((((.(((...........))).)))))))))................((((((((((((((((((((----............)))))))..)))))))...)))))).. ( -36.40, z-score = -3.56, R) >droYak2.chr2L 11346633 110 + 22324452 ACGAUGGUAUCAGUAAUCUGCGGCC----AGAUAUAGUCAGAGAUAAAACCCAUAUCGUGAGUGGGGCAACGAAU----UACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAU ....((((..(((....)))..)))----).........................((((((((((((((((((((----............)))))))..)))))))...)))))).. ( -31.20, z-score = -2.25, R) >droEre2.scaffold_4929 9290085 110 + 26641161 ACGAUGGUAUCAGCAAUCUGCGGCC----AGAUAUAGUUCAUGAUAAAGCCCAUAUCGGGAAUGGGGCAACGAAU----UACAGUGCAACUGUUUGUUUAGCCCCACAAAUCGCGAAU ....((((....((.....)).)))----)......((((.((((....(((.....)))..(((((((((((..----.((((.....)))))))))..))))))...)))).)))) ( -31.00, z-score = -1.32, R) >droAna3.scaffold_12916 10344747 94 + 16180835 AUAAUAACCCCACUUAUCUUUACCC----AGAUAUAGUCGCAGAUAAAGUUCACCUCGGGGAUGGGCCAACCAGU----UACAGUG--GUUGGGGAUUGCACCU-------------- .......((((((((((((......----))))).)))....((......)).....)))).....(((((((..----.....))--)))))((......)).-------------- ( -21.70, z-score = 0.74, R) >consensus ACGACGGUAUCAGUAAUCUGCGGCC____AGAUAUAGUCAGAGAUAAAACCCAUAUCGCGAGUGGGGCAACGAAU____UACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAU ..(((.(((((...................))))).)))................((((((((((((((((((((................)))))))..)))))))...)))))).. (-17.25 = -19.83 + 2.59)

| Location | 8,698,052 – 8,698,172 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.48 |
| Shannon entropy | 0.37093 |
| G+C content | 0.43902 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -17.35 |
| Energy contribution | -19.36 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8698052 120 + 23011544 CAGAGAUAAAAUCCAUAUCGCGAGUGGGGCAACGAAUGAAUUACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAUCGAUGGUCCCAUAAUGAGGAGAGAUCUACUACAUAAAUUG .((((((....(((...(((((((((((((((((((..(............)..))))))..)))))))...)))))).((.(((....)))...)))))...)))).)).......... ( -36.10, z-score = -3.68, R) >droSim1.chr2L 8483297 116 + 22036055 CAGAGAUAAAACCCAUAUCGCGAGUGGGGCAACGAAU----UACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAUCGAUGGACACAUAAUGAGGAGAGAUAUACUACAUGAAUUG ...........(((((.((((((((((((((((((((----............)))))))..)))))))...))))))....(((....))).))).))..................... ( -30.50, z-score = -3.00, R) >droSec1.super_3 4179804 116 + 7220098 CAGAGAUAAAAUCCAUAUCGCGAGUGGGGCAACGAAU----UACAGUGCAAUUGUUUGUUAAGCCCCACAAAUCGCGAAUCGAUGGACACAUAAUGAGGAGAGAUCUACUACAUGAAUUG .((((((....(((...((((((((((((((((((((----............)))))))..)))))))...)))))).((.(((....)))...)))))...)))).)).......... ( -35.00, z-score = -3.97, R) >droYak2.chr2L 11346667 113 + 22324452 CAGAGAUAAAACCCAUAUCGUGAGUGGGGCAACGAAU----UACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAUCGAUGAACACAUAAUGAGGAGAGAU---CUACAUGAAUUG ((.((((....(((((.((((((((((((((((((((----............)))))))..)))))))...))))))....(((....))).))).))....))---))...))..... ( -30.70, z-score = -3.01, R) >droEre2.scaffold_4929 9290119 113 + 26641161 UCAUGAUAAAGCCCAUAUCGGGAAUGGGGCAACGAAU----UACAGUGCAACUGUUUGUUUAGCCCCACAAAUCGCGAAUCGAUGAACACAUAAUGAGGAGAGAU---CUACAUGAAUUG (((((......(((((.(((.((.(((((((((((..----.((((.....)))))))))..))))))....)).)))....(((....))).))).))((....---)).))))).... ( -27.90, z-score = -1.50, R) >droAna3.scaffold_12916 10344781 94 + 16180835 CGCAGAUAAAGUUCACCUCGGGGAUGGGCCAACCAGU----UACAGUG---------GUUGGG-------GAUUGCACCUCGAUGGGCACAUAAUGAGGAGAGAUCUGGUAGAG------ ..(((((....(((.(((((...(((..(((((((..----.....))---------))))).-------...(((.((.....))))))))..))))).))))))))......------ ( -29.90, z-score = -1.48, R) >consensus CAGAGAUAAAACCCAUAUCGCGAGUGGGGCAACGAAU____UACAGUGCAAUUGUUUGUUUAGCCCCACAAAUCGCGAAUCGAUGGACACAUAAUGAGGAGAGAUCUACUACAUGAAUUG ............((...((((((((((((((((((((................)))))))..)))))))...)))))).((.(((....)))...))))..................... (-17.35 = -19.36 + 2.00)

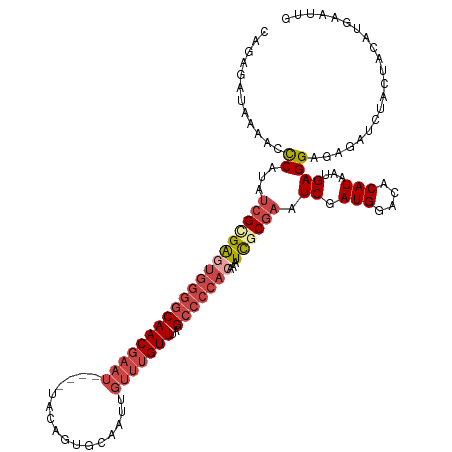
| Location | 8,698,052 – 8,698,172 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Shannon entropy | 0.37093 |
| G+C content | 0.43902 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -12.55 |
| Energy contribution | -14.08 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
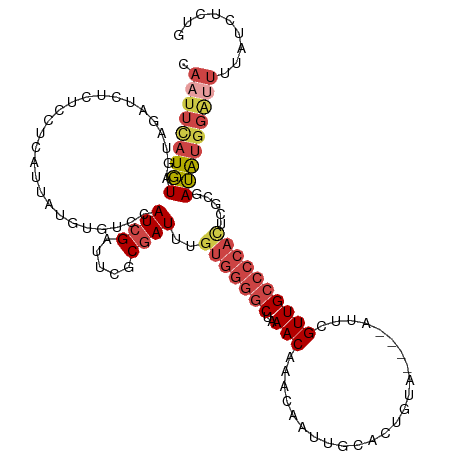
>dm3.chr2L 8698052 120 - 23011544 CAAUUUAUGUAGUAGAUCUCUCCUCAUUAUGGGACCAUCGAUUCGCGAUUUGUGGGGCUAAACAAACAAUUGCACUGUAAUUCAUUCGUUGCCCCACUCGCGAUAUGGAUUUUAUCUCUG .........(((.((((...((((......))))((((....((((((...(((((((.........((((((...))))))........))))))))))))).)))).....))))))) ( -32.83, z-score = -2.73, R) >droSim1.chr2L 8483297 116 - 22036055 CAAUUCAUGUAGUAUAUCUCUCCUCAUUAUGUGUCCAUCGAUUCGCGAUUUGUGGGGCUAAACAAACAAUUGCACUGUA----AUUCGUUGCCCCACUCGCGAUAUGGGUUUUAUCUCUG .....((((((((............))))))))(((((....((((((...(((((((......(((((((((...)))----))).)))))))))))))))).)))))........... ( -30.60, z-score = -2.98, R) >droSec1.super_3 4179804 116 - 7220098 CAAUUCAUGUAGUAGAUCUCUCCUCAUUAUGUGUCCAUCGAUUCGCGAUUUGUGGGGCUUAACAAACAAUUGCACUGUA----AUUCGUUGCCCCACUCGCGAUAUGGAUUUUAUCUCUG .....((((((((((.......)).))))))))(((((....((((((...(((((((......(((((((((...)))----))).)))))))))))))))).)))))........... ( -33.10, z-score = -3.50, R) >droYak2.chr2L 11346667 113 - 22324452 CAAUUCAUGUAG---AUCUCUCCUCAUUAUGUGUUCAUCGAUUCGCGAUUUGUGGGGCUAAACAAACAAUUGCACUGUA----AUUCGUUGCCCCACUCACGAUAUGGGUUUUAUCUCUG ..........((---((....((.(((..((((...((((.....))))..(((((((......(((((((((...)))----))).)))))))))).))))..)))))....))))... ( -25.50, z-score = -1.37, R) >droEre2.scaffold_4929 9290119 113 - 26641161 CAAUUCAUGUAG---AUCUCUCCUCAUUAUGUGUUCAUCGAUUCGCGAUUUGUGGGGCUAAACAAACAGUUGCACUGUA----AUUCGUUGCCCCAUUCCCGAUAUGGGCUUUAUCAUGA ....((((((((---(.(((...((...........((((.....))))..(((((((.......((((.....)))).----.......)))))))....))...))).)))).))))) ( -26.66, z-score = -0.74, R) >droAna3.scaffold_12916 10344781 94 - 16180835 ------CUCUACCAGAUCUCUCCUCAUUAUGUGCCCAUCGAGGUGCAAUC-------CCCAAC---------CACUGUA----ACUGGUUGGCCCAUCCCCGAGGUGAACUUUAUCUGCG ------......(((((.((.((((....((..((......))..))...-------.(((((---------((.....----..))))))).........)))).)).....))))).. ( -26.30, z-score = -2.75, R) >consensus CAAUUCAUGUAGUAGAUCUCUCCUCAUUAUGUGUCCAUCGAUUCGCGAUUUGUGGGGCUAAACAAACAAUUGCACUGUA____AUUCGUUGCCCCACUCGCGAUAUGGAUUUUAUCUCUG .(((((((((..........................((((.....))))..(((((((................................))))))).....)))))))))......... (-12.55 = -14.08 + 1.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:37 2011