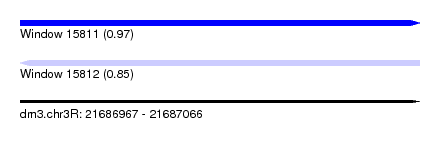
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,686,967 – 21,687,066 |
| Length | 99 |
| Max. P | 0.973487 |

| Location | 21,686,967 – 21,687,066 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.75 |
| Shannon entropy | 0.45241 |
| G+C content | 0.43979 |
| Mean single sequence MFE | -31.11 |
| Consensus MFE | -15.61 |
| Energy contribution | -15.51 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
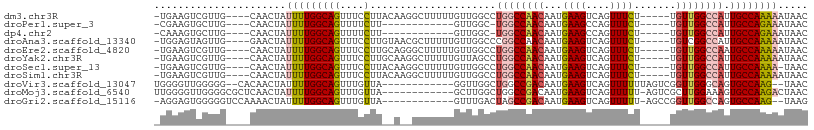
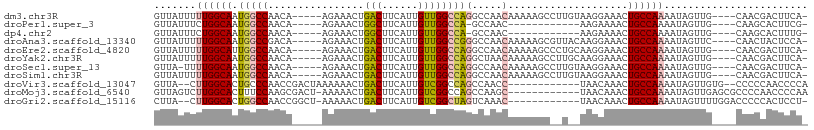
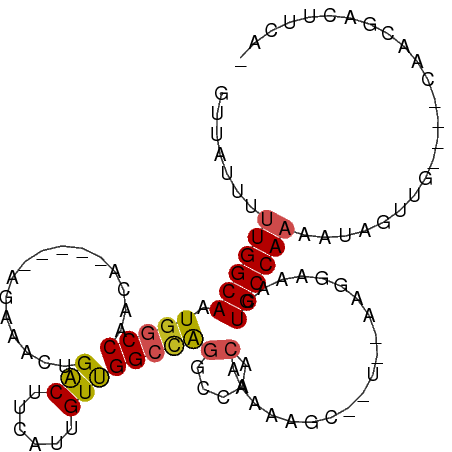
>dm3.chr3R 21686967 99 + 27905053 -UGAAGUCGUUG----CAACUAUUUUGGCAGUUUCCUUACAAGGCUUUUUGUUGGCCUGGCCAACAAUGAAGUCAGUUUCU-----UGUUGGCCAUUGCCAAAAAUAAC -...........----......((((((((((.........(((((.......)))))(((((((((.((((....)))))-----))))))))))))))))))..... ( -34.70, z-score = -3.67, R) >droPer1.super_3 1203042 86 - 7375914 -CGAAGUGCUUG----CAACUAUUUUGGCAGUUUUCUU------------GUUGGC-UGGCCAACAAUGAAGCCAGUUUCU-----UGUUGGCCAUUGCCAGAAAUAAC -.(((.((((..----..........)))).)))....------------.(((((-((((((((((.((((....)))))-----)))))))))..)))))....... ( -29.20, z-score = -2.50, R) >dp4.chr2 7393124 86 - 30794189 -CAAAGUGCUUG----CAACUAUUUUGGCAGUUUUCUU------------GUUGGC-UGGCCAACAAUGAAGCCAGUUUCU-----UGUUGGCCAUUGCCAGAAAUAAC -.(((.((((..----..........)))).)))....------------.(((((-((((((((((.((((....)))))-----)))))))))..)))))....... ( -28.80, z-score = -2.29, R) >droAna3.scaffold_13340 12238857 99 - 23697760 -UGGAGUAGUUG----GAACUAUUUUGGCAGUUUCCUUGUAACGCUUUUUGUUGGCCCGGCCAACAAUGAAGUCAGUUUCU-----UGUCGGCCAUUGCCAAAAAUAAC -.((((((((..----..))))))))((((((..((..(.((((((((((((((((...)))))))).)))))..))).).-----....))..))))))......... ( -26.90, z-score = -1.09, R) >droEre2.scaffold_4820 4107736 99 - 10470090 -UGAAGUCGUUG----CAACUAUUUUGGCAGUUUCCUUGCAGGGCUUUUUGUUGGCCUGGCCAACAAUGAAGUCAGUUUCU-----UGUUGGCCAAUGCCAAAAAUAAC -...........----......((((((((...........(((((.......)))))(((((((((.((((....)))))-----))))))))..))))))))..... ( -33.40, z-score = -2.44, R) >droYak2.chr3R 4037417 99 - 28832112 -UGAAGUCGUUG----CAACUAUUUUGGCAGUUUCCUUGCAAGGCUUUUUGUUAGCCUGGCCAACAAUGAAGUCAGUUUCU-----UGUUGGCCAUUGCCAAAAAUAAC -...........----......((((((((((.........(((((.......)))))(((((((((.((((....)))))-----))))))))))))))))))..... ( -34.70, z-score = -3.67, R) >droSec1.super_13 465826 98 + 2104621 -UGAAGUCGUUG----CAACUAUUUUGGCAGUUUCCUUACAAGGCUUUUUGUUGGCCUGGCCAACAAUGAAGUCAGUUUCU-----UGUUGGCCAUUGCCAAAA-UAAC -...........----....((((((((((((.........(((((.......)))))(((((((((.((((....)))))-----))))))))))))))))))-)).. ( -36.90, z-score = -4.35, R) >droSim1.chr3R 21515954 99 + 27517382 -UGAAGUCGUUG----CAACUAUUUUGGCAGUUUCCUUACAAGGCUUUUUGUUGGCCUGGCCAACAAUGAAGUCAGUUUCU-----UGUUGGCCAUUGCCAAAAAUAAC -...........----......((((((((((.........(((((.......)))))(((((((((.((((....)))))-----))))))))))))))))))..... ( -34.70, z-score = -3.67, R) >droVir3.scaffold_13047 16096628 93 + 19223366 UGGGGUUGGGGG--CACAACUAUUUUGGCAGUUUGUUA------------GGUUGGCUGGCCGACAAUGAAGUCAGUUUUUUAGUCGGUUGGGCAGUGCCAAG--UAAC ....((((..((--(((......((((((.....))))------------)).((.(..((((((..(((((......)))))))))))..).)))))))...--)))) ( -29.30, z-score = -2.10, R) >droMoj3.scaffold_6540 14898581 96 + 34148556 UUGGGGUUGGGGCGCUCAACUAUUUUGGCAGUUUGUUA------------GCUUGGCUGGCCGACAAUGAAGUCAGUUUUU-AGUCGCUUGGAAAGUGCCAAGACUAAC (((((((((((...))))))).((((((((.(((.(.(------------((..(((((((.(((......))).))...)-))))))).).))).)))))))))))). ( -30.30, z-score = -1.26, R) >droGri2.scaffold_15116 941421 93 + 1808639 -AGGAGUGGGGGUCCAAAACUAUUUUGGCAGUUUGUUA------------GUUUGACUAGCCGACAAUGAAGUCAGUUUUU-AGCCGGUUGGCCAGUGCCAAG--UAAG -.........(((..((((((.....(((((((.....------------....)))).)))(((......))))))))).-.)))(.(((((....))))).--)... ( -23.30, z-score = -0.18, R) >consensus _UGAAGUCGUUG____CAACUAUUUUGGCAGUUUCCUU__A__GCUUUUUGUUGGCCUGGCCAACAAUGAAGUCAGUUUCU_____UGUUGGCCAUUGCCAAAAAUAAC ......................(((((((((....).....................((((((((...((((....)))).......)))))))).))))))))..... (-15.61 = -15.51 + -0.10)
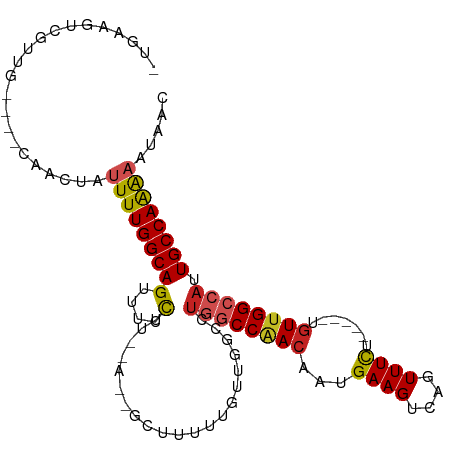
| Location | 21,686,967 – 21,687,066 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.75 |
| Shannon entropy | 0.45241 |
| G+C content | 0.43979 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -10.01 |
| Energy contribution | -10.59 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21686967 99 - 27905053 GUUAUUUUUGGCAAUGGCCAACA-----AGAAACUGACUUCAUUGUUGGCCAGGCCAACAAAAAGCCUUGUAAGGAAACUGCCAAAAUAGUUG----CAACGACUUCA- .....(((((((..(((((((((-----((((......))).))))))))))(((.........))).....((....))))))))).(((((----...)))))...- ( -32.30, z-score = -3.28, R) >droPer1.super_3 1203042 86 + 7375914 GUUAUUUCUGGCAAUGGCCAACA-----AGAAACUGGCUUCAUUGUUGGCCA-GCCAAC------------AAGAAAACUGCCAAAAUAGUUG----CAAGCACUUCG- (((.....((((..(((((((((-----((((......))).))))))))))-))))..------------.....(((((......))))).----..)))......- ( -25.00, z-score = -1.98, R) >dp4.chr2 7393124 86 + 30794189 GUUAUUUCUGGCAAUGGCCAACA-----AGAAACUGGCUUCAUUGUUGGCCA-GCCAAC------------AAGAAAACUGCCAAAAUAGUUG----CAAGCACUUUG- (((.....((((..(((((((((-----((((......))).))))))))))-))))..------------.....(((((......))))).----..)))......- ( -25.00, z-score = -1.66, R) >droAna3.scaffold_13340 12238857 99 + 23697760 GUUAUUUUUGGCAAUGGCCGACA-----AGAAACUGACUUCAUUGUUGGCCGGGCCAACAAAAAGCGUUACAAGGAAACUGCCAAAAUAGUUC----CAACUACUCCA- (((.((((((((..(((((((((-----((((......))).)))))))))).))))).))).))).......((((.(((......))))))----)..........- ( -29.90, z-score = -2.84, R) >droEre2.scaffold_4820 4107736 99 + 10470090 GUUAUUUUUGGCAUUGGCCAACA-----AGAAACUGACUUCAUUGUUGGCCAGGCCAACAAAAAGCCCUGCAAGGAAACUGCCAAAAUAGUUG----CAACGACUUCA- (((.((((((((.((((((((((-----((((......))).)))))))))))))))).))).)))...(((.(....))))......(((((----...)))))...- ( -32.20, z-score = -2.89, R) >droYak2.chr3R 4037417 99 + 28832112 GUUAUUUUUGGCAAUGGCCAACA-----AGAAACUGACUUCAUUGUUGGCCAGGCUAACAAAAAGCCUUGCAAGGAAACUGCCAAAAUAGUUG----CAACGACUUCA- .....(((((((..(((((((((-----((((......))).))))))))))((((.......)))).....((....))))))))).(((((----...)))))...- ( -34.40, z-score = -3.96, R) >droSec1.super_13 465826 98 - 2104621 GUUA-UUUUGGCAAUGGCCAACA-----AGAAACUGACUUCAUUGUUGGCCAGGCCAACAAAAAGCCUUGUAAGGAAACUGCCAAAAUAGUUG----CAACGACUUCA- ..((-(((((((..(((((((((-----((((......))).))))))))))(((.........))).....((....)))))))))))((((----...))))....- ( -34.40, z-score = -4.02, R) >droSim1.chr3R 21515954 99 - 27517382 GUUAUUUUUGGCAAUGGCCAACA-----AGAAACUGACUUCAUUGUUGGCCAGGCCAACAAAAAGCCUUGUAAGGAAACUGCCAAAAUAGUUG----CAACGACUUCA- .....(((((((..(((((((((-----((((......))).))))))))))(((.........))).....((....))))))))).(((((----...)))))...- ( -32.30, z-score = -3.28, R) >droVir3.scaffold_13047 16096628 93 - 19223366 GUUA--CUUGGCACUGCCCAACCGACUAAAAAACUGACUUCAUUGUCGGCCAGCCAACC------------UAACAAACUGCCAAAAUAGUUGUG--CCCCCAACCCCA ....--...(((((.((....(((((..................)))))...)).....------------.....(((((......))))))))--)).......... ( -16.47, z-score = -1.24, R) >droMoj3.scaffold_6540 14898581 96 - 34148556 GUUAGUCUUGGCACUUUCCAAGCGACU-AAAAACUGACUUCAUUGUCGGCCAGCCAAGC------------UAACAAACUGCCAAAAUAGUUGAGCGCCCCAACCCCAA (((((.((((((....((.....))..-.....(((((......)))))...)))))))------------)))).....(((((.....))).))............. ( -22.70, z-score = -1.96, R) >droGri2.scaffold_15116 941421 93 - 1808639 CUUA--CUUGGCACUGGCCAACCGGCU-AAAAACUGACUUCAUUGUCGGCUAGUCAAAC------------UAACAAACUGCCAAAAUAGUUUUGGACCCCCACUCCU- ....--.((((((.(((((....))))-)....(((((......)))))..........------------........))))))........(((....))).....- ( -20.90, z-score = -1.68, R) >consensus GUUAUUUUUGGCAAUGGCCAACA_____AGAAACUGACUUCAUUGUUGGCCAGGCCAACAAAAAGC__U__AAGGAAACUGCCAAAAUAGUUG____CAACGACUUCA_ .......((((((.(((((.((......................)).)))))(.....)....................))))))........................ (-10.01 = -10.59 + 0.57)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:14 2011