| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,676,640 – 21,676,749 |
| Length | 109 |
| Max. P | 0.596171 |

| Location | 21,676,640 – 21,676,749 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 57.26 |
| Shannon entropy | 0.90373 |
| G+C content | 0.40685 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -8.11 |
| Energy contribution | -8.30 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.43 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
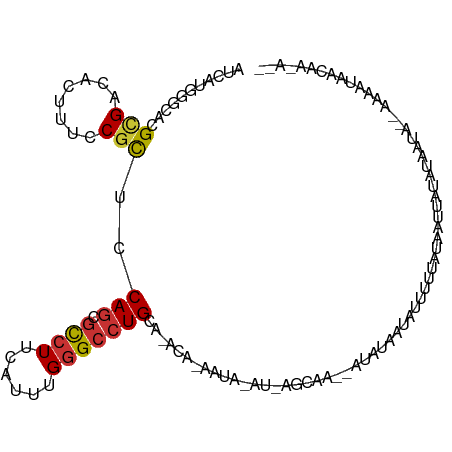
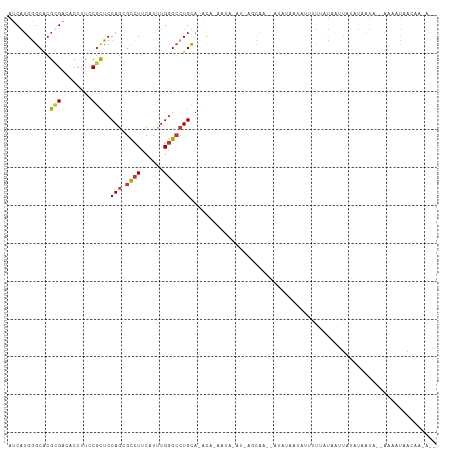
>dm3.chr3R 21676640 109 + 27905053 AUCAUGGGCACGCGACACUUUCCGCUCCAGGGCCUUCAUUUGGGCCUGCACAUACAAAAUGGAUGCAAGCAUAUACGAUUUUAUUUAAAAUAUUGUU--AAAAUUUCAACU-- ...(((.((..(((........)))....((((((......)))))))).)))......((((....((((......(((((....)))))..))))--.....))))...-- ( -19.00, z-score = 0.45, R) >droWil1.scaffold_181130 2294498 112 - 16660200 AUCGAGAGCCCGUGACACCUUACGCUCCAGCGUCUUCAUUUGGGGCUGCA-ACGGGAUACAUGGAUACAAAAAGGUUAUAGCCAUAUGUUUUUCAAAGAUGGUUAAAAGCAUU (((.(...(((((..((((((((((....))))........)))).))..-))))).....).))).......(.((.(((((((.((.....))...))))))).)).)... ( -26.10, z-score = -0.05, R) >droPer1.super_3 1190864 109 - 7375914 GUCGUGGGCACGCGUCACUUUACGCUCCAGCGCCUUCAUUUGGGCCUGCA-ACAAAAUUGAUUAGCAA--AUACCCCGCUUUUACAAUUUUCCAAUAACAAUGCAACAAAAU- ...((..(((.((((......))))..(((.((((......)))))))..-..((((((((..(((..--.......)))..).)))))))..........))).)).....- ( -21.60, z-score = -0.34, R) >dp4.chr2 7380973 103 - 30794189 GUCGUGGGCACGCGUCACUUUACGCUCCAGCGCCUUCAUUUGGGCCUGCA-ACAAAAUUGAUCAGCAA--AUACCACGCUUUUACAAUUUUCCAACA------CAACAAAAU- ((.(((.((..((((......))))...((.((((......)))))))).-..((((((((..(((..--.......)))..).)))))))....))------).)).....- ( -21.10, z-score = -1.11, R) >droAna3.scaffold_13340 12227094 102 - 23697760 GUCAUGAGCGCGUGAAACCUUCCUUUCCAACGCUUUCAUUUGUGCCUGC---CAUGGUGUAUUAUUAGUCAACAUAUAUGAUUUUCCUUCU--UAUUGUAAUCUAAU------ ...(((((.((((((((......))))..)))).))))).((..((...---...))..))..(((((...(((.(((.((.......)).--))))))...)))))------ ( -16.80, z-score = -1.03, R) >droEre2.scaffold_4820 4097117 96 - 10470090 AUCAUGGGCACGCGACACUUUCCGCUCCAGGGCCUUCAUUUGGGCCUGUAUAU------------CAA--AUUUACGAUUUUUAUUAUAAUUUAGUAC-AAAACUUCAAUU-- .((.((((...(((........)))))))((((((......))))))......------------...--......))(((.(((((.....))))).-))).........-- ( -16.60, z-score = -0.74, R) >droYak2.chr3R 4026646 97 - 28832112 AUCGUGGGCACGCGAUACCUUCCGCUCCAGCGCCUUCAUUUGGGCCUGCAUAU------------AGAGAAUAUAUAUGUAUCAUUAUUAUAUAUUG--AAAAUUGCAAUU-- ((((((....)))))).......((..(((.((((......))))))).....------------....((((((((.(((....))))))))))).--......))....-- ( -21.10, z-score = -0.16, R) >droSec1.super_13 456066 106 + 2104621 AUCAUGGGCACGCGAAACUUUCCGCUCCAGGGCCUUCAUUUGGGCCUGCA--CAUACAAAAUGGAUA-CAAGCAUAUUCGAUUUUCUUUAUAUUGUUAAAAUUUUAACU---- ((((((.((..(((........)))....((((((......)))))))).--))).......(((((-......))))))))............(((((....))))).---- ( -20.40, z-score = -0.50, R) >consensus AUCAUGGGCACGCGACACUUUCCGCUCCAGCGCCUUCAUUUGGGCCUGCA_ACA_AAUA_AU_AGCAA__AUAUAAUAUUUUUAUAAUUAUAUAAUA__AAAAUAACAA_A__ ...........(((........)))..(((.((((......)))))))................................................................. ( -8.11 = -8.30 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:11 2011