| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,589,353 – 21,589,449 |
| Length | 96 |
| Max. P | 0.951287 |

| Location | 21,589,353 – 21,589,449 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 64.23 |
| Shannon entropy | 0.68069 |
| G+C content | 0.49319 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -10.14 |
| Energy contribution | -10.20 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21589353 96 + 27905053 ACCGCGGCGCAUUAGCAGAGGUUUUCCCACCAAACGAAACAGUUGUGGAUGCUGCUGGAAAAGCGCUGUUAAGC----------CACUGCUAACCGCUUAACAGUG----- .(((((((((....))..........((((..(((......)))))))..))))).)).....(((((((((((----------...........)))))))))))----- ( -31.20, z-score = -1.25, R) >droSim1.chr3R 21417143 95 + 27517382 ACCGCGGCGCAUUAGCAGAGGUUUUCCCACCAG-CGAAACAGUUGUGGAUGCUGCUGGAAAAGCGCUGUUAAGC----------CACUGCUAACCGCUUAACAGUG----- .(((((((((....))..........((((.((-(......)))))))..))))).)).....(((((((((((----------...........)))))))))))----- ( -32.80, z-score = -1.24, R) >droSec1.super_13 377509 96 + 2104621 ACCGCGGCGCAUUAGCAGAGGUUUUUCCCACCAGCGAAACAUUUGUGGAUGCUGCUGGAAAAGCGCUGUUAAGC----------CACUGCUAACCGCUUAACAGUG----- ...((((....(((((((..((((((((...(((((..((....))...)))))..))))))))(((....)))----------..))))))))))).........----- ( -33.50, z-score = -1.89, R) >droYak2.chr3R 3944688 106 - 28832112 ACCGCGGCGCAUUAGCAGAGGUUUUUCCCUCCAGCAAAACAGUUGUGGAUGCUGCUGGAAAAGCGCUGUUAAGCGCUGUUAAGCCACUGCUAACCGCUUAACAGUG----- ..((((((((.(((((((..((((((((...(((((..((....))...)))))..)))))))).))))))))))))(((((((...........))))))).)))----- ( -41.80, z-score = -2.68, R) >droEre2.scaffold_4820 4012012 95 - 10470090 ACCGCGGCGCAUUAGCAGAGGUUUUCCC-UCCAGCAAAACAGUUGUGGAUGCCGCUGGAACAGCGCUGUUAGGC----------CACUGCUAAGCGCUUAACAGCG----- .(((((((((....)).((((.....))-))((((......)))).....))))).)).....(((((((((((----------..((....)).)))))))))))----- ( -36.80, z-score = -1.60, R) >droAna3.scaffold_13340 12147141 91 - 23697760 ACCGCGAAGCAUUACCAGAGAUUUUCCGCAC-----UAACAAUUGUGGAUGCUGCUGUUAAGUUACUGUUAGCC----------AGCUUUCAAGAACUUAACAGUG----- .......((((.............((((((.-----.......))))))))))(((((((((((..((..((..----------..))..))..))))))))))).----- ( -24.91, z-score = -2.24, R) >dp4.chr2 7284298 72 - 30794189 ------------------------GCCGCAGCACAUUAACAGUUGUGGAUGCUGCUGUUAAAGCGCUGCUAAUC----------CUCUGCCAAAAACUUAACAUUG----- ------------------------...(((((.(.((((((((.((....)).)))))))).).))))).....----------......................----- ( -20.30, z-score = -1.94, R) >droPer1.super_3 1094075 72 - 7375914 ------------------------GCCGCAGCGCAUUAACAGUUGUGGAUGCUGCUGUUAAAGCGCUGCUAAUC----------CUCUGCCAAAAACUUAACAUUG----- ------------------------...(((((((.((((((((.((....)).)))))))).))))))).....----------......................----- ( -25.70, z-score = -3.71, R) >droWil1.scaffold_181130 2203866 87 - 16660200 CCCACAACGCAUUAACAGU--UUUGUCUUUUC---------AUUGUGGAUGCCAUUAACAGAGUGCUGUUAUUA-------------CACUAAAGCGAUAACAGCGGCAGA .((((((.......((...--...))......---------.)))))).(((((((.....)))((((((((..-------------(......)..)))))))))))).. ( -22.44, z-score = -2.02, R) >consensus ACCGCGGCGCAUUAGCAGAGGUUUUCCCCACCA_CGAAACAGUUGUGGAUGCUGCUGGAAAAGCGCUGUUAAGC__________CACUGCUAACCGCUUAACAGUG_____ .......................................((((.((....)).))))......((((((((((.......................))))))))))..... (-10.14 = -10.20 + 0.06)

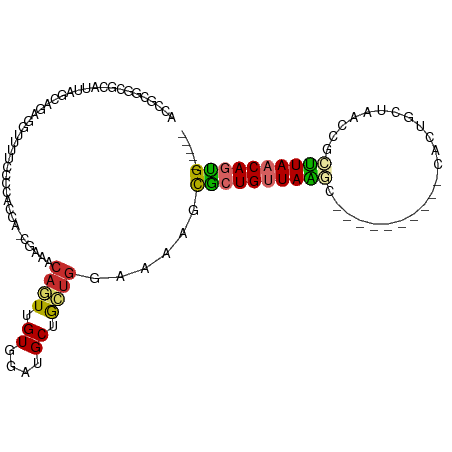
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:06 2011