
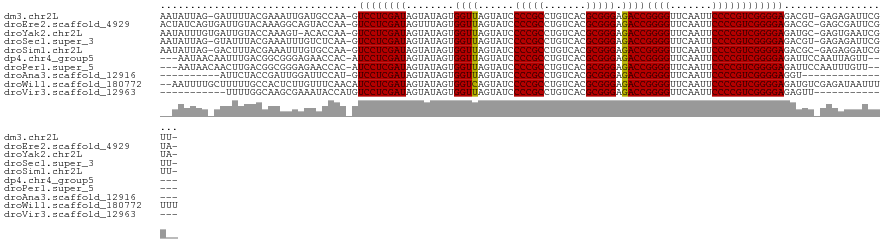
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,679,084 – 8,679,203 |
| Length | 119 |
| Max. P | 0.939565 |

| Location | 8,679,084 – 8,679,203 |
|---|---|
| Length | 119 |
| Sequences | 10 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Shannon entropy | 0.48647 |
| G+C content | 0.51662 |
| Mean single sequence MFE | -37.96 |
| Consensus MFE | -30.20 |
| Energy contribution | -30.11 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
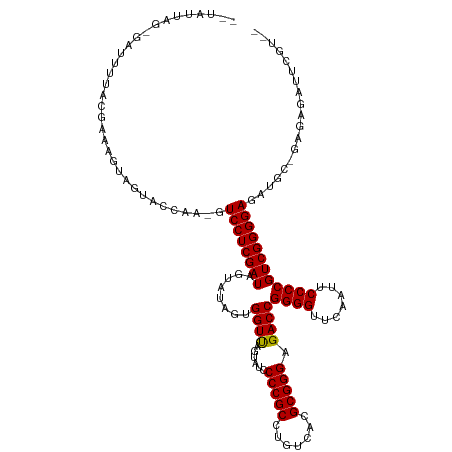
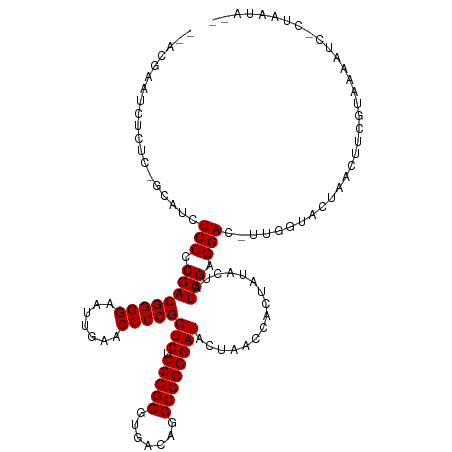
>dm3.chr2L 8679084 119 + 23011544 AAUAUUAG-GAUUUUACGAAAUUGAUGCCAA-GUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGU-GAGAGAUUCGUU- .......(-(((((((((...(((....)))-.((((((((........((((......(((((.......))))).))))((((.......))))))))))))..)))-))..)))))...- ( -36.60, z-score = -0.63, R) >droEre2.scaffold_4929 9270644 120 + 26641161 ACUAUCAGUGAUUGUACAAAGGCAGUACCAA-GUCCUCGAUAGUUUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGC-GAGCGAUUCGUA- ((((((((.((((((((.......))))..)-))))).))))))..(((.(((.((.(((((.((((.....)))).(((.((((.......)))))))))))).))..-.))).)))....- ( -41.20, z-score = -1.22, R) >droYak2.chr2L 11323033 119 + 22324452 AAUAUUUGUGAUUGUACCAAAGU-ACACCAA-GUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUGC-GAGUGAAUCGUA- ..(((((((...(((((....))-)))....-.((((((((........((((......(((((.......))))).))))((((.......))))))))))))...))-))))).......- ( -37.90, z-score = -1.24, R) >droSec1.super_3 4158539 119 + 7220098 AAUAUUAG-GUAUUUACGAAAUUUGUCUCAA-GUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGU-GAGAGAUUCGUU- ........-......(((((...(.(((((.-.((((((((........((((......(((((.......))))).))))((((.......))))))))))))....)-)))).)))))).- ( -39.00, z-score = -1.50, R) >droSim1.chr2L 8461858 119 + 22036055 AAUAUUAG-GACUUUACGAAAUUUGUGCCAA-GUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGC-GAGAGGAUCGUU- ..((((((-((((((((((...)))))..))-))))).))))............((.(((((.((((.....)))).(((.((((.......)))))))))))).))((-((.....)))).- ( -40.50, z-score = -1.04, R) >dp4.chr4_group5 1451792 114 - 2436548 ---AAUAACAAUUUGACGGCGGGAGAACCAC-AUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUUCCAAUUAGUU----- ---..........(((((((((((.((((((-((.((....)).)).))))))....).)))))).))))..(((....)))((..((...(((((...)))))))..))........----- ( -40.00, z-score = -1.56, R) >droPer1.super_5 1423524 114 - 6813705 ---AAUAACAACUUGACGGCGGGAGAACCAC-AUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUUCCAAUUUGUU----- ---...(((((..(((((((((((.((((((-((.((....)).)).))))))....).)))))).))))..(((....)))((..((...(((((...)))))))..))...)))))----- ( -41.00, z-score = -1.53, R) >droAna3.scaffold_12916 10321829 96 + 16180835 ----------AUUCUACCGAUUGGAUUCCAU-GUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGGU---------------- ----------.....(((.((((((((((..-(((...((((.((.......)).))))(((((.......))))).))).))))))))))(((((...))))))))---------------- ( -33.60, z-score = -0.36, R) >droWil1.scaffold_180772 8095441 121 + 8906247 --AAUUUUGCUUUUUGCCACUCUUGUUUCAACAUCCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUGUCGAGAUAAUUUUUU --......((.....)).....(((((((.(((((((((((........((((......(((((.......))))).))))((((.......))))))))))))..))).)))))))...... ( -39.00, z-score = -2.13, R) >droVir3.scaffold_12963 14508309 98 - 20206255 -----------UUUUGGCAAGCGAAAUACCAUGUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAGUU-------------- -----------(((((.....))))).....(.((((((((........((((......(((((.......))))).))))((((.......)))))))))))).)...-------------- ( -30.80, z-score = -0.26, R) >consensus __UAUUAG_GAUUUUACGAAAGUAGUACCAA_GUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUGC_GAGAGAUUCGU__ .................................((((((((........((((......(((((.......))))).))))((((.......))))))))))))................... (-30.20 = -30.11 + -0.09)
| Location | 8,679,084 – 8,679,203 |
|---|---|
| Length | 119 |
| Sequences | 10 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 76.95 |
| Shannon entropy | 0.48647 |
| G+C content | 0.51662 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.33 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
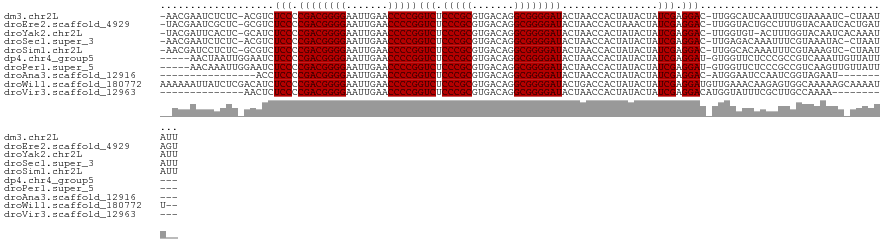
>dm3.chr2L 8679084 119 - 23011544 -AACGAAUCUCUC-ACGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAC-UUGGCAUCAAUUUCGUAAAAUC-CUAAUAUU -.(((((......-.((..(((.((((((((.......)))))(((.(((((.......))))))))................))).))).-.))........)))))......-........ ( -33.26, z-score = -0.85, R) >droEre2.scaffold_4929 9270644 120 - 26641161 -UACGAAUCGCUC-GCGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAAACUAUCGAGGAC-UUGGUACUGCCUUUGUACAAUCACUGAUAGU -..(((.....))-).((.((((((.(((((.......)))))(((........)))...)))))).))..........(((((((..((.-...((((.......))))..))..))))))) ( -37.00, z-score = -0.74, R) >droYak2.chr2L 11323033 119 - 22324452 -UACGAUUCACUC-GCAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAC-UUGGUGU-ACUUUGGUACAAUCACAAAUAUU -...((((.....-.((..(((.((((((((.......)))))(((.(((((.......))))))))................))).))).-.))..((-((....))))))))......... ( -34.70, z-score = -0.99, R) >droSec1.super_3 4158539 119 - 7220098 -AACGAAUCUCUC-ACGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAC-UUGAGACAAAUUUCGUAAAUAC-CUAAUAUU -.(((((..((((-(.((((...((((((((.......)))))(((.(((((.......))))))))................))).))))-.))))).....)))))......-........ ( -37.00, z-score = -2.29, R) >droSim1.chr2L 8461858 119 - 22036055 -AACGAUCCUCUC-GCGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAC-UUGGCACAAAUUUCGUAAAGUC-CUAAUAUU -..(((.....))-).((.((((((.(((((.......)))))(((........)))...)))))).)).................(((((-((.((.........)).)))))-))...... ( -35.30, z-score = -0.76, R) >dp4.chr4_group5 1451792 114 + 2436548 -----AACUAAUUGGAAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAU-GUGGUUCUCCCGCCGUCAAAUUGUUAUU--- -----........((.....)).((.((((..((((.....))))..))))))((((.(((((((.....((((((....((....))...-)))))).)))))))))))..........--- ( -37.30, z-score = -1.05, R) >droPer1.super_5 1423524 114 + 6813705 -----AACAAAUUGGAAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAU-GUGGUUCUCCCGCCGUCAAGUUGUUAUU--- -----(((((...((.....)).((.((((..((((.....))))..))))))((((.(((((((.....((((((....((....))...-)))))).)))))))))))..)))))...--- ( -39.10, z-score = -1.23, R) >droAna3.scaffold_12916 10321829 96 - 16180835 ----------------ACCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAC-AUGGAAUCCAAUCGGUAGAAU---------- ----------------...((((((.(((((.......)))))(((........)))...))))))..............((((((((((.-......)))..)))))))...---------- ( -32.90, z-score = -0.90, R) >droWil1.scaffold_180772 8095441 121 - 8906247 AAAAAAUUAUCUCGACAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUGACCACUAUACUAUCGAGGAUGUUGAAACAAGAGUGGCAAAAAGCAAAAUU-- ...........(((((((((...(((.((((.......))))((((((((((.......)))))).....)))).........))).)))))))))..........((.....))......-- ( -35.60, z-score = -1.70, R) >droVir3.scaffold_12963 14508309 98 + 20206255 --------------AACUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACAUGGUAUUUCGCUUGCCAAAA----------- --------------.....(((.((((((((.......)))))(((.(((((.......))))))))................))).)))..(((((.......)))))...----------- ( -32.00, z-score = -1.38, R) >consensus __ACGAAUCUCUC_GCAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAC_UUGGUACUAACUUCGUAAAAUC_CUAAUA__ ...................(((.((((((((.......)))))(((.(((((.......))))))))................))).)))................................. (-27.33 = -27.33 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:35 2011