
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,587,725 – 21,587,820 |
| Length | 95 |
| Max. P | 0.537706 |

| Location | 21,587,725 – 21,587,820 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.50 |
| Shannon entropy | 0.60353 |
| G+C content | 0.44072 |
| Mean single sequence MFE | -22.56 |
| Consensus MFE | -10.55 |
| Energy contribution | -10.41 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21587725 95 + 27905053 CGGCGUAACGGUCCACUUGAAAUGGG-CAUAAUUUAUGAUUUGCGCGUGUAAAUCAUUUUUCAAGCACCACC------CC-GCGAUGCUUGUGAAUGACAGAG------------- ..((((((..(((((.......))))-)............)))))).......((((((..((((((.(...------..-..).)))))).)))))).....------------- ( -21.24, z-score = 0.40, R) >droSim1.chr3R 21415503 95 + 27517382 CGGCGUAACGGGCCACUUGAAAUGGG-CAUAAUUUAUGAUUUGCGCGUGUAAAUCAUUUUUCAAGCACCACC------CC-GCGAUGCUUGUGACUGACAGAG------------- .(((((..((((...(((((((....-........(((((((((....))))))))).)))))))......)------))-)..)))))..............------------- ( -23.52, z-score = -0.03, R) >droSec1.super_13 375870 95 + 2104621 CGGCGUAACGGGCCACUUGAAAUGGG-CAUAAUUUAUGAUUUGCGCGUGUAAAUCAUUUUUCAAGCACCACC------CC-GCGAUGCUUGUGACUGACAGAG------------- .(((((..((((...(((((((....-........(((((((((....))))))))).)))))))......)------))-)..)))))..............------------- ( -23.52, z-score = -0.03, R) >droYak2.chr3R 3942900 95 - 28832112 CGGCAUAACGGGCCACUUGAAAUGGG-CAUAAUUUAUGAUUUGCGCGUGUAAAUCAUUUUUCAAGCACCACC------CC-GCGAUGCUUGUGAAUGACAGAA------------- .(((((..((((...(((((((....-........(((((((((....))))))))).)))))))......)------))-)..)))))..............------------- ( -23.92, z-score = -0.32, R) >droEre2.scaffold_4820 4010318 83 - 10470090 ------------CCACUUGAAAUGGG-CAUAAUUUAUGAUUUGCGCGUGUAAAUCAUUUUUCAAGCACCAGC------CC-GCGAUGCUUGUGAAUGACAGGA------------- ------------...((((..(((.(-((............))).))).....((((((..((((((...(.------..-.)..)))))).)))))))))).------------- ( -19.90, z-score = -0.42, R) >droAna3.scaffold_13340 12145659 84 - 23697760 ------------CCACUUGAAGUAGG-CAUAAUUUAUGAUUUACGCGUGUAAAUCAUUUUUCAAGCACCACC------CCCGCCAUAUUUGUGCCUGACAGUU------------- ------------.........(((((-(((((...(((((((((....))))))))).......((......------...)).....)))))))).))....------------- ( -18.30, z-score = -2.38, R) >dp4.chr2 7282106 110 - 30794189 UGUCCUUUCGGGCCACUUGAAAUGGGACAUAAUUUAUGAUUUACGCGUGUAAAUCAUUUUUCAAGCACCACC------CCCAGCCAGGUUGCGAGUGAUACUUGCUCGUGACAGUU ((((((((((((...))))))..))))))......(((((((((....)))))))))...............------.........((..((((..(...)..))))..)).... ( -29.70, z-score = -1.30, R) >droPer1.super_3 1091810 110 - 7375914 UGUCGUUUCGGGCCACUUGAAAUGGGACAUAAUUUAUGAUUUACGCGUGUAAAUCAUUUUUCAAGCCCCACC------CCCAGCCGGGUUGCGAGUGAUACUUGCUCGUGACAGUU (((((...(((((((((((....(((.........(((((((((....))))))))).........)))(((------(......))))..))))))......))))))))))... ( -32.37, z-score = -1.50, R) >droWil1.scaffold_181130 2202559 79 - 16660200 ----------GGCCACUUGAGAGGC--CAUAAUUUAUGAUUUAUGUGUGUAAAUCAUAUUUCAAACGCACUG-------CUUCUCUGUUCGCACACUU------------------ ----------.((.((..(((((((--.......((((((((((....)))))))))).............)-------)))))).))..))......------------------ ( -17.75, z-score = -1.67, R) >droVir3.scaffold_12855 7019676 87 + 10161210 ------------CCACUUGAACUG---CAUAAUUUAUGAUUUGCGUGUGUAAAUCAUUUUUCAAGCACCCGCCA-UGAAACUGCGUGUUUGCCGUUGACAGAC------------- ------------.........(((---........(((((((((....)))))))))...(((((((..(((((-......)).)))..)))..)))))))..------------- ( -17.00, z-score = -0.24, R) >droMoj3.scaffold_6540 8547237 80 - 34148556 ------------CCACUUCAAGUG---CGUAAUUUAUGAUUUACGUGUGUAAAUCAUUUUUCAAGUGG--------AGCUCUCUCUGUUUGCCGUUGACAGAG------------- ------------((((((.((((.---....))))(((((((((....))))))))).....))))))--------......(((((((.......)))))))------------- ( -20.70, z-score = -2.37, R) >droGri2.scaffold_15074 1951546 88 - 7742996 ------------GCACUUGAAGUG---CAUAAUUUAUGAUUUACGUGUGUAAAUCAUUUGUGAAGCACCCGCAAGCAGAGUUUCGUGUUUGCCGUUGACAGAU------------- ------------((((.....)))---)........((((((((....))))))))(((((..(((....((((((((.....).))))))).))).))))).------------- ( -22.80, z-score = -1.43, R) >consensus __________GGCCACUUGAAAUGGG_CAUAAUUUAUGAUUUACGCGUGUAAAUCAUUUUUCAAGCACCACC______CC_GCCAUGCUUGCGAAUGACAGAG_____________ ...............(((((((.............(((((((((....))))))))).)))))))................................................... (-10.55 = -10.41 + -0.14)

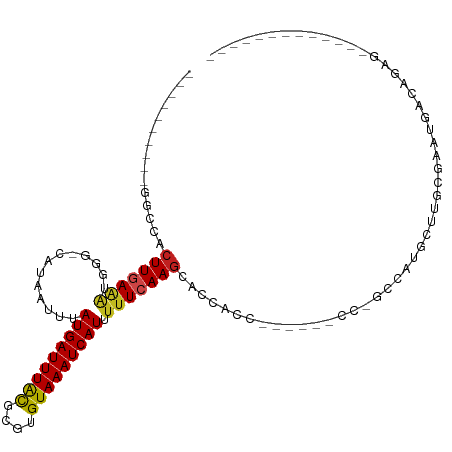
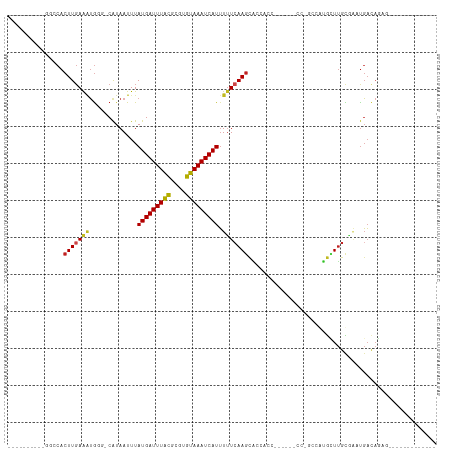
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:05 2011