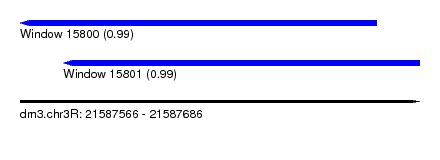
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,587,566 – 21,587,686 |
| Length | 120 |
| Max. P | 0.989320 |

| Location | 21,587,566 – 21,587,673 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 67.49 |
| Shannon entropy | 0.52589 |
| G+C content | 0.42842 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -13.32 |
| Energy contribution | -15.92 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
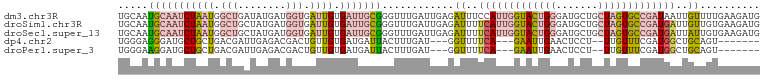
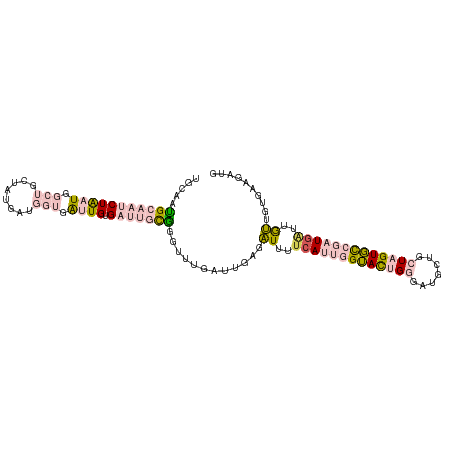
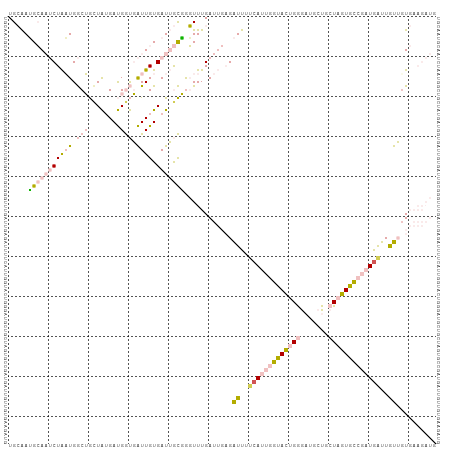
>dm3.chr3R 21587566 107 - 27905053 UGCAAUGCAAUCUAAUGGCUGAUAUGAUGGUGAUUGUGAUUGCGGGUUUGAUUGAGAUUUCCAUUGGUACUGGGAUGCUGCUAGUGCCGAUAAUUGUUUUGAAGAUG .....(((((((((((.(((........))).)))).))))))).......(..((((....(((((((((((.......)))))))))))....))))..)..... ( -30.60, z-score = -2.51, R) >droSim1.chr3R 21415344 107 - 27517382 UGCAAUGCAAUCUAAUGGCUGCUAUGAUGGUGAUUGUGAUUGCGGGUUUGAUUGAGAUUUUCAUUGGUACUGGGAUGCUGCUAGUGCCGAUGAUUGUUGUGAAGAUG .(((((((((((((((.(((........))).)))).)))))).................(((((((((((((.......)))))))))))))..)))))....... ( -34.40, z-score = -3.70, R) >droSec1.super_13 375711 107 - 2104621 UGCAAUGCAAUCUAAUGGCUGCUAUGAUGGUGAUUGUGAUUGCGGGUUUGAUUGAGAUUUUCAUUGGUACUGGGAUGCUGCUAGUGCCGAUGAUUAUUGUGAAGAUG .(((((((((((((((.(((........))).)))).)))))).................(((((((((((((.......)))))))))))))..)))))....... ( -34.10, z-score = -3.63, R) >dp4.chr2 7281968 92 + 30794189 UGGGAGGGAUGCUGCUGACGAUUGAGACGACUGUUGUGAUGAUUACUUUGAU---GGUUUUCA---GAAUUGAACUCCU--UUGUUUCGAUGGCUGCAGU------- ..........(((((.(.(.((((((((((..(((((..(((..(((.....---)))..)))---..))..)))....--)))))))))).)).)))))------- ( -23.40, z-score = -1.32, R) >droPer1.super_3 1091670 92 + 7375914 UGGGAAGGAUGCUGCUGACGAUUGAGACGACUGUUGUGAUGAUUACUUUGAU---GGUUUUCA---GAAUUGAACUCCU--UUGUUUCGAUGGCUGCAGU------- ..........(((((.(.(.((((((((((..(((((..(((..(((.....---)))..)))---..))..)))....--)))))))))).)).)))))------- ( -23.40, z-score = -1.52, R) >consensus UGCAAUGCAAUCUAAUGGCUGCUAUGAUGGUGAUUGUGAUUGCGGGUUUGAUUGAGAUUUUCAUUGGUACUGGGAUGCUGCUAGUGCCGAUGAUUGUUGUGAAGAUG .....(((((((((((.(((........))).)))).)))))))............((..(((((((((((((.......)))))))))))))..)).......... (-13.32 = -15.92 + 2.60)
| Location | 21,587,579 – 21,587,686 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.55 |
| Shannon entropy | 0.55235 |
| G+C content | 0.42948 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -17.20 |
| Energy contribution | -17.45 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21587579 107 - 27905053 UGACGAUGGUGAUUGCAAUGCAAUCUAAUGGCUGAUAUGAUGGUGAUUGUGAUUGCGGGUUUGAUUGAGAUUU-------------CCAUUGGUACUGGGAUGCUGCUAGUGCCGAUAAU ....(((((.(((..((((((((((((((.(((........))).)))).)))))).......))))..))).-------------)))))((((((((.......))))))))...... ( -31.61, z-score = -2.51, R) >droSim1.chr3R 21415357 107 - 27517382 UGACGAUGGCGAUUGCAAUGCAAUCUAAUGGCUGCUAUGAUGGUGAUUGUGAUUGCGGGUUUGAUUGAGAUUU-------------UCAUUGGUACUGGGAUGCUGCUAGUGCCGAUGAU ....(((..(((((....(((((((((((.(((........))).)))).))))))).....)))))..))).-------------(((((((((((((.......))))))))))))). ( -35.10, z-score = -3.28, R) >droSec1.super_13 375724 107 - 2104621 UGACGAUGGCGAUUGCAAUGCAAUCUAAUGGCUGCUAUGAUGGUGAUUGUGAUUGCGGGUUUGAUUGAGAUUU-------------UCAUUGGUACUGGGAUGCUGCUAGUGCCGAUGAU ....(((..(((((....(((((((((((.(((........))).)))).))))))).....)))))..))).-------------(((((((((((((.......))))))))))))). ( -35.10, z-score = -3.28, R) >droEre2.scaffold_4820 4010160 120 + 10470090 UGACGAUGGCGAUUGCAAUGCCAUCUAAUGGCAGAUAUGAUGGCGAUUGUGACUGCGGGUUUGAUAGAGAUUUGGAUUGGGUAUUGGUAUUGGUAUUGGAAUGCUGCUAGUGCCAAUGAU ...((((((((.(..((((((((((.............)))))).))))..).))).((((..(.......)..))))...))))).((((((((((((.......)))))))))))).. ( -35.92, z-score = -2.33, R) >dp4.chr2 7281974 99 + 30794189 AAAUAAUGAUGAUUGGGAGGGAUGCUGCUGACGAUUGAGACGACUGUUGUGAUGAUUACUUUGAUGG-------------------UUUUCAGAAUUGAACUCCU--UUGUUUCGAUGGC ............(..(.((.....)).)..)(.((((((((((..(((((..(((..(((.....))-------------------)..)))..))..)))....--)))))))))).). ( -18.00, z-score = -0.03, R) >droPer1.super_3 1091676 99 + 7375914 GAAUAUUGAUGAUUGGGAAGGAUGCUGCUGACGAUUGAGACGACUGUUGUGAUGAUUACUUUGAUGG-------------------UUUUCAGAAUUGAACUCCU--UUGUUUCGAUGGC ...((((((.....((((..(((.(((..(((.((..((((....)))((((...)))).)..)).)-------------------))..))).)))....))))--.....)))))).. ( -16.50, z-score = 0.51, R) >consensus UGACGAUGGCGAUUGCAAUGCAAUCUAAUGGCUGAUAUGAUGGCGAUUGUGAUUGCGGGUUUGAUUGAGAUUU_____________UCAUUGGUACUGGAAUGCUGCUAGUGCCGAUGAU ........(((((((((((((.(((.............))).))).))))))))))..............................(((((((((((((.......))))))))))))). (-17.20 = -17.45 + 0.26)

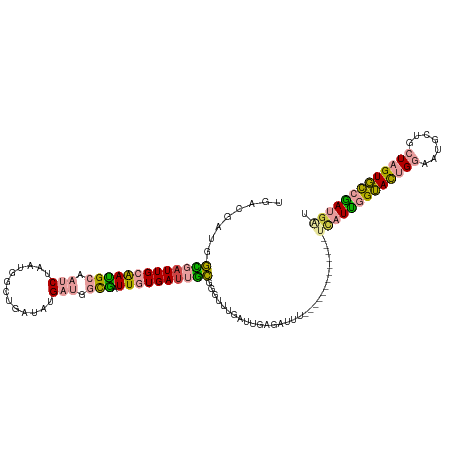
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:04 2011