
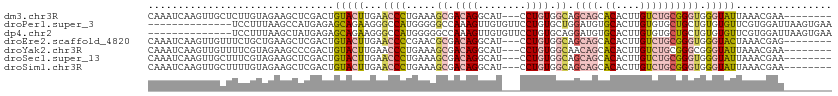
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,574,258 – 21,574,361 |
| Length | 103 |
| Max. P | 0.522658 |

| Location | 21,574,258 – 21,574,361 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 67.09 |
| Shannon entropy | 0.57655 |
| G+C content | 0.50212 |
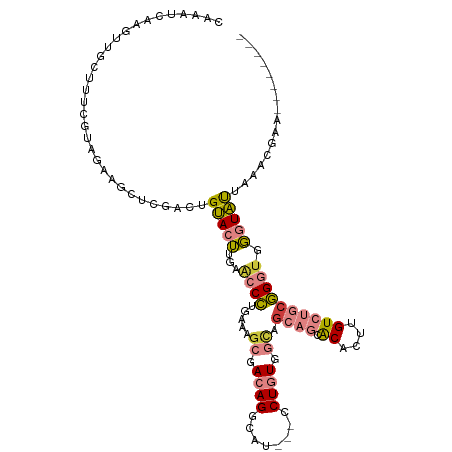
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -14.94 |
| Energy contribution | -15.84 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21574258 103 + 27905053 CAAAUCAAGUUGCUCUUGUAGAAGCUCGACUGUACUUGAACCCUGAAAGCGACAGGCAU---CCUGUGGCAGCAGCACACUUGUCUGCGGGUGGGUAUUAAACGAA-------- ....((.(((((..(((....)))..)))))(((((...((((.....((.(((((...---))))).)).((((.((....)))))))))).))))).....)).-------- ( -31.20, z-score = -0.78, R) >droPer1.super_3 1076776 100 - 7375914 --------------UCCUUUAAGCCAUGAGAGCAGAAGGGCCAUGGGGGCCAAAGUUGUGUUCCUGUGCUGGAUGUGCACUUGUGUGCUGCUGUGUGUUCGUGGAUUAAGUGAA --------------((.(((((.(((((((.((((...(((.((((((((((....)).)))))))))))......((((....))))..))))...))))))).))))).)). ( -33.90, z-score = -1.44, R) >dp4.chr2 7266984 100 - 30794189 --------------UCCUUUAAGCUAUGAGAGCAGAAGGGCCAUGGGGGCCAAAGUUGUGUUCCUGUGCAGGAUGUGCACUUGUGUGCUGCUGUGUGUUCGUGGAUUAAGUGAA --------------((.(((((.(((((((.((((...((((((((((((((....)).))))))(((((.....)))))..))).))).))))...))))))).))))).)). ( -32.10, z-score = -1.04, R) >droEre2.scaffold_4820 3996233 103 - 10470090 CAAAUCAAGUUGUUUCUGCUGAAGCUCGACUGUACUUGAACCCCGAACGCGACAGGCAU---CCUGUGGCAGCAGCACACUUGUCUGCGGGUGGGUACUAAACGAG-------- ...........(((((....)))))(((...(((((...((((.....((.(((((...---))))).)).((((.((....)))))))))).)))))....))).-------- ( -34.20, z-score = -1.29, R) >droYak2.chr3R 3928616 103 - 28832112 CAAAUCAAGUUGUUUUCGUAGAAGCCCGACUGUACUUGAACCCUGAAAGCGACAGGCAU---CCUGUGGCAACAGCACACUUGUCUGCGGGCGGGUAUUAAACGAA-------- ..............(((((..(((((((.(((((..(((...(((...((.(((((...---))))).))..))).....)))..))))).))))).))..)))))-------- ( -27.60, z-score = -0.07, R) >droSec1.super_13 362058 103 + 2104621 CAAAUCAAGUUGCUUUCGUAGAAGCUCGACUGUACUUGAACCCUGAAAGCGACAGGCAU---CCUGUGGCAGCAGCACACUUGUCUGCGGGUGGGUAUUAAACGAA-------- ....((((((.((..(((........)))..))))))))((((.....((.(((((...---))))).)).((((.((....))))))))))..............-------- ( -32.80, z-score = -1.34, R) >droSim1.chr3R 21401526 103 + 27517382 CAAAUCAAGUUGCUUUUGUAGAAGCUCGACUGUACUUGAACCCUGAAAGCGACAGGCAU---CCUGUGGCAGCAGCACACUUGUCUGCGGGUGGGUAUUAAACGAA-------- ...........(((((....)))))(((...(((((...((((.....((.(((((...---))))).)).((((.((....)))))))))).)))))....))).-------- ( -32.90, z-score = -1.41, R) >consensus CAAAUCAAGUUGCUUUCGUAGAAGCUCGACUGUACUUGAACCCUGAAAGCGACAGGCAU___CCUGUGGCAGCAGCACACUUGUCUGCGGGUGGGUAUUAAACGAA________ ...............................(((((...((((.....((.(((((......))))).)).((((.((....)))))))))).)))))................ (-14.94 = -15.84 + 0.90)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:44:00 2011