
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,572,522 – 21,572,591 |
| Length | 69 |
| Max. P | 0.717029 |

| Location | 21,572,522 – 21,572,591 |
|---|---|
| Length | 69 |
| Sequences | 12 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 77.26 |
| Shannon entropy | 0.46116 |
| G+C content | 0.50826 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -12.21 |
| Energy contribution | -11.79 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.717029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21572522 69 + 27905053 CAAAAGCUGCCAAAGCUGGCA-GGGCGCUGAGUUAUGCCCAACAUGGCCAUAAUUCAUGUGGCUUAGGCG-------- .....(((((((....))))(-((.((((((((((((.((.....)).))))))))).))).))).))).-------- ( -27.20, z-score = -1.80, R) >droAna3.scaffold_13340 12131627 68 - 23697760 CAAAAGCUGCCAAAGCUGGCA-GGACGCUGAGUUAUGCCCGACUUGGCCAUAAUUUACGUGGCUUAAAC--------- ...(((((((((....)))).-..(((.(((((((((.((.....)).)))))))))))))))))....--------- ( -21.90, z-score = -1.98, R) >droEre2.scaffold_4820 3994471 69 - 10470090 CAAAAGCUGCCAAAGCUGGCA-GGGCGCUGAGUUAUGCCCAACUUGGCCAUAAUUCAUGUGGCUUAGGCG-------- .....(((((((....))))(-((.((((((((((((.((.....)).))))))))).))).))).))).-------- ( -27.20, z-score = -1.73, R) >droYak2.chr3R 3926828 69 - 28832112 CAAAAGCUGCCAAAGCUGGCA-GGGCGCUGAGUUAUGCCCAACUUGGCCAUAAUUCAUGUGGCUUAGGCG-------- .....(((((((....))))(-((.((((((((((((.((.....)).))))))))).))).))).))).-------- ( -27.20, z-score = -1.73, R) >droSec1.super_13 360317 69 + 2104621 CAAAAGCUGCCAAAGCUGGCA-GGGCGCUGAGUUAUGCCCAACUUGGCCAUAAUUCAUGUAGCUUAGGCG-------- .....(((((((....)))).-(((((((((((((((.((.....)).))))))))).)).)))).))).-------- ( -24.60, z-score = -1.18, R) >droSim1.chr3R 21399790 69 + 27517382 CAAAAGCUGCCAAAGCUGGCA-GGGCGCUGAGUUAUGCCCAACUUGGCCAUAAUUCAUGUGGCUUAGGCG-------- .....(((((((....))))(-((.((((((((((((.((.....)).))))))))).))).))).))).-------- ( -27.20, z-score = -1.73, R) >droWil1.scaffold_181130 2184874 63 - 16660200 CAAAAGCUGCCAAAGCUGACU---GCUGUGAGUUAUGCCCAACUUGCUCGUAAUUUAUAUGCCACA------------ ....((((.....))))....---(((((((((((((..(.....)..))))))))))).))....------------ ( -13.30, z-score = -0.85, R) >droPer1.super_3 1075516 66 - 7375914 CAAAAGCUGCCAAAGCUGCUUUGGUCCCUGAGUUAUGCCCAACUUGGUCGUAAUUUAUGUGGCUUA------------ ...((((((((((((...)))))))...(((((((((.((.....)).)))))))))...))))).------------ ( -19.30, z-score = -1.98, R) >dp4.chr2 7265725 66 - 30794189 CAAAAGCUGCCAAAGCUGCUUUGGUCCCUGAGUUAUGCCCAACUUGGUCGUAAUUUAUGUGGCUUA------------ ...((((((((((((...)))))))...(((((((((.((.....)).)))))))))...))))).------------ ( -19.30, z-score = -1.98, R) >droMoj3.scaffold_6540 8529716 65 - 34148556 CAAAAGCUGCCAAAGCUGGCA---GCGCUGAGUUAUGCCCAACUUGGCUGUAAUUUAUAUGCCAGGCG---------- .....(((((((....)))))---))((((((((......)))))(((((((...)))).))).))).---------- ( -22.60, z-score = -1.36, R) >droVir3.scaffold_12855 6999419 65 + 10161210 CAAAAGCUGCCAAAGCUGGCA---GCGCUGAGUUAUGCUCAACUUGGCUGUAAUUUAUAUGCCAGGCG---------- .....(((((((....)))))---))..((((.....)))).((((((((((...)))).))))))..---------- ( -25.00, z-score = -2.48, R) >triCas2.ChLG5 13000728 76 - 18847211 CCAAAGCCGUUAUCGCUGGUA-GUGCGGC-ACUUGGGCGGAAUGUGGCCCCAGUUCAAACGCCACACAAUCUCAGGUG .....((((((((.....)))-..)))))-(((((((.....((((((............))))))....))))))). ( -23.30, z-score = -0.06, R) >consensus CAAAAGCUGCCAAAGCUGGCA_GGGCGCUGAGUUAUGCCCAACUUGGCCAUAAUUCAUGUGGCUUAGGC_________ ....((((.....))))...........(((((((((.((.....)).)))))))))..................... (-12.21 = -11.79 + -0.42)

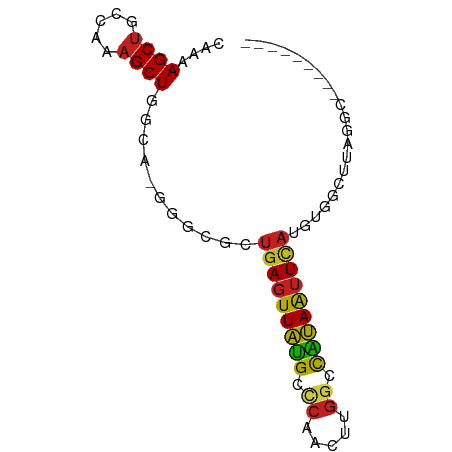

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:59 2011