
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,569,516 – 21,569,626 |
| Length | 110 |
| Max. P | 0.563966 |

| Location | 21,569,516 – 21,569,626 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.87 |
| Shannon entropy | 0.43031 |
| G+C content | 0.38973 |
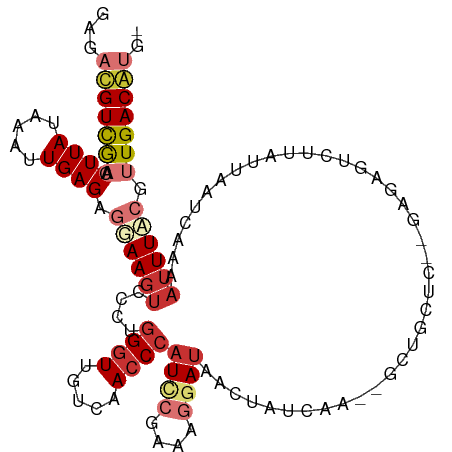
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -13.92 |
| Energy contribution | -15.38 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.563966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
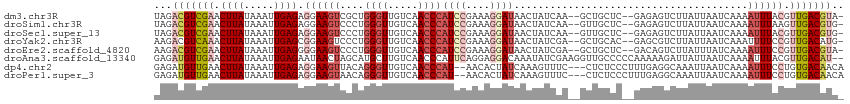
>dm3.chr3R 21569516 110 + 27905053 UAGACGUCGAACUUAUAAAUUGAGAGGAAGUCGCUGGGUUGUCAACCCAUCCGAAAGGAUAACUAUCAA--GCUGCUC--GAGAGUCUUAUUAAUCAAAAUUUACGUUGACGUA- ...(((((((.........((((.(((((((.(((((((.....))))((((....))))........)--)).)))(--....))))).....))))........))))))).- ( -28.03, z-score = -1.54, R) >droSim1.chr3R 21396355 110 + 27517382 UAGACGUCGAACUUAUAAAUUGAGAGGAAGUCCCUGGGUUGUCAACCCAUCCGAAAGGAUAACUAUCAA--GUUGCUC--GAGAGUCUUAUUAAUCAAAAUUUAAGUUGACGUG- ...(((((.((((((....((((....(((.(.((((((.....))))((((....))))((((....)--)))....--.)).).))).....))))....))))))))))).- ( -29.50, z-score = -2.49, R) >droSec1.super_13 357352 110 + 2104621 UAGACGUCGAACUUAUAAAUUGAGAGGAAGUCCCUGGGUUGUCAACCCAUCCGAAAGGAUAACUAUCAA--GUUGCUC--GAGAGUCUUAUUAAUCAAAAUUUACGUUGACGUG- ...(((((((.....((((((..((..(((.(.((((((.....))))((((....))))((((....)--)))....--.)).).))).....))..))))))..))))))).- ( -25.80, z-score = -1.06, R) >droYak2.chr3R 3923842 110 - 28832112 AAGACGUCAAACUUAUAAAUUGAGCGGAAGUCCCUGGGUUGUCAACCCAUCCGAAAGGAUAACUAUCGA--GCUGCAC--GAGCGUCUUAUUAAUCAAAUUUUCCGUUGACAUG- .....(((((.((((.....))))((((((.....((((.....))))((((....)))).......((--(.(((..--..))).)))...........)))))))))))...- ( -27.60, z-score = -1.71, R) >droEre2.scaffold_4820 3991579 110 - 10470090 AAGACGUCGAACUUAUAAAUUGAGGGGAAGUCCCUGGGUUGUCAACCCAUCCGAAAGGAUAACUAUCGA--GCUGCUC--GACAGUCUUAUUUAUCAAAAUUUCCGUUGACGUA- ...(((((((.((((.....)))).((((((....((((.....))))((((....)))).(((.((((--(...)))--)).))).............)))))).))))))).- ( -33.50, z-score = -2.52, R) >droAna3.scaffold_13340 12128610 113 - 23697760 GAGAUGUUGAACUUAUAAAUUGAGAAUAACUAGCAUGCUUGUCAACCCAUUCAGGAGGACAAAUAUCGAAGGUUGCCCCCAAAAAGAUUAUUAAUCAAAAUUUACGUUGACAU-- (((((((((..((((.....))))......)))))).)))((((((.......((.((.(((....(....)))))).)).....(((.....))).........))))))..-- ( -17.70, z-score = -0.11, R) >dp4.chr2 7262817 110 - 30794189 GAGAUGUUGAACUUAUAAAUUGAGAGGAAGUUACAGGGUUGUCAACCCAU--AACACUAUCAAAGUUUC---CUCUCCCUUUGAGGCAAAUUAAUCAAAAUUUCCUGUGACAACA ....(((((.((...((((..((((((((((((..((((.....)))).)--)))(((.....))))))---)))))..))))(((.(((((......))))))))))..))))) ( -26.70, z-score = -1.88, R) >droPer1.super_3 1072573 110 - 7375914 GAGAUGUUGAACUUAUAAAUUGAGAGGAAGUAACAGGGUUGUCAACCCAU--AACACUAUCAAAGUUUC---CUCUCCCUUUGAGGCAAAUUAAUCAAAAUUUCCUGUGACAACA ....(((((.((...((((..((((((((......((((.....))))..--...(((.....))))))---)))))..))))(((.(((((......))))))))))..))))) ( -25.20, z-score = -1.46, R) >consensus GAGACGUCGAACUUAUAAAUUGAGAGGAAGUCCCUGGGUUGUCAACCCAUCCGAAAGGAUAACUAUCAA__GCUGCUC__GAGAGUCUUAUUAAUCAAAAUUUACGUUGACAUG_ ...(((((((.((((.....)))).((((((....((((.....))))((((....)))).......................................)))))).))))))).. (-13.92 = -15.38 + 1.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:59 2011