| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,558,685 – 21,558,750 |
| Length | 65 |
| Max. P | 0.580739 |

| Location | 21,558,685 – 21,558,750 |
|---|---|
| Length | 65 |
| Sequences | 5 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 63.85 |
| Shannon entropy | 0.67353 |
| G+C content | 0.37544 |
| Mean single sequence MFE | -13.37 |
| Consensus MFE | -6.72 |
| Energy contribution | -5.92 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.68 |
| Mean z-score | -0.54 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
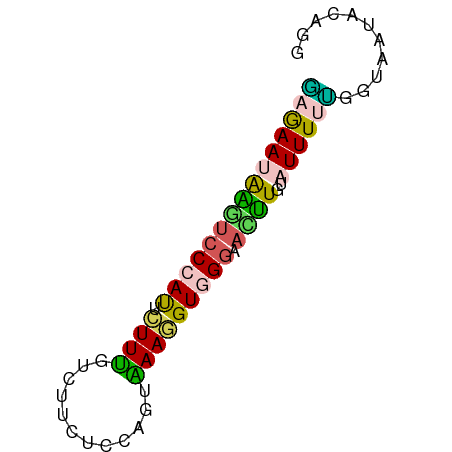
>dm3.chr3R 21558685 65 + 27905053 UUAAAAAGAUCCCAUUCUUUGGAAUCACCAAAGAAGGUGGGAAUCUGUAUUUUAGGCAGGGUUGG .........(((((((((((((.....)))))))..)))))).(((((.......)))))..... ( -20.00, z-score = -2.44, R) >droSim1.chr2h_random 793290 65 + 3178526 GAGAAAAAGUCCAACUCUUUUUCUUCUUCAGUAAAGGUGGGAACUUGUAUUUUUGGUAAUACUUU (((((((((.......))))))))).........((((....))))(((((......)))))... ( -10.00, z-score = 0.40, R) >droSec1.super_80 44630 65 + 135508 AAGAAUAAGUCCCACUAUUUGACUUCUCCAGUAAAGGUAGGAACUUGUAUUUUUGGUUAUACAGG .........(((.(((.....(((.....)))...))).))).(((((((........))))))) ( -9.70, z-score = 0.78, R) >droEre2.scaffold_4784 616793 65 - 25762168 GAGAAUAAUUCCCAUUCUUUUCCUUCUCCAGUAAAGGUGGGAACUUGUAUUUUUGGUAAUACAGG ........(((((((.((((.(........).)))))))))))((((((((......)))))))) ( -15.80, z-score = -1.40, R) >triCas2.singleUn_1817 15298 58 - 22356 CUGAAUGCUAACCAUUUUUCGUAUGCACCGUCGAAAGUGGGCAAGUCAAUUUUGGGUA------- ...........(((.........(((.(((.(....))))))).........)))...------- ( -11.37, z-score = -0.05, R) >consensus GAGAAUAAGUCCCAUUCUUUGUCUUCUCCAGUAAAGGUGGGAACUUGUAUUUUUGGUAAUACAGG (((((((((((((((.((((............))))))))).))))..))))))........... ( -6.72 = -5.92 + -0.80)


| Location | 21,558,685 – 21,558,750 |
|---|---|
| Length | 65 |
| Sequences | 5 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 63.85 |
| Shannon entropy | 0.67353 |
| G+C content | 0.37544 |
| Mean single sequence MFE | -11.90 |
| Consensus MFE | -4.76 |
| Energy contribution | -4.20 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
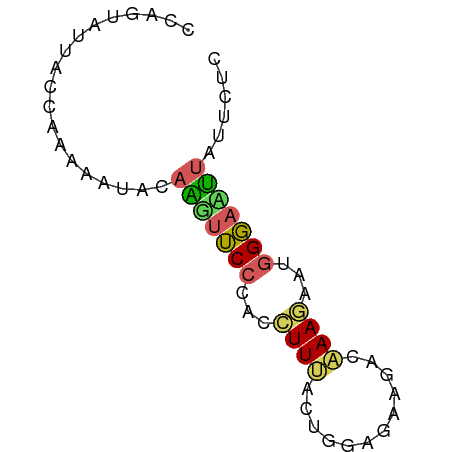
>dm3.chr3R 21558685 65 - 27905053 CCAACCCUGCCUAAAAUACAGAUUCCCACCUUCUUUGGUGAUUCCAAAGAAUGGGAUCUUUUUAA ...........(((((...((((.((((..((((((((.....))))))))))))))))))))). ( -17.80, z-score = -3.38, R) >droSim1.chr2h_random 793290 65 - 3178526 AAAGUAUUACCAAAAAUACAAGUUCCCACCUUUACUGAAGAAGAAAAAGAGUUGGACUUUUUCUC ...(((((......)))))((((.((.((((((.((.....))..)))).)).))))))...... ( -8.70, z-score = -0.08, R) >droSec1.super_80 44630 65 - 135508 CCUGUAUAACCAAAAAUACAAGUUCCUACCUUUACUGGAGAAGUCAAAUAGUGGGACUUAUUCUU ...((((........))))((((.(((((..(((((.....)))..))..)))))))))...... ( -10.50, z-score = -0.07, R) >droEre2.scaffold_4784 616793 65 + 25762168 CCUGUAUUACCAAAAAUACAAGUUCCCACCUUUACUGGAGAAGGAAAAGAAUGGGAAUUAUUCUC ..((((((......))))))(((((((((((((......))))).......))))))))...... ( -15.21, z-score = -1.68, R) >triCas2.singleUn_1817 15298 58 + 22356 -------UACCCAAAAUUGACUUGCCCACUUUCGACGGUGCAUACGAAAAAUGGUUAGCAUUCAG -------..........(((..((((((.(((((..........)))))..)))...))).))). ( -7.30, z-score = 0.26, R) >consensus CCAGUAUUACCAAAAAUACAAGUUCCCACCUUUACUGGAGAAGACAAAGAAUGGGAAUUAUUCUC ...................(((((((...((((............))))...)))))))...... ( -4.76 = -4.20 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:56 2011