| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,551,010 – 21,551,119 |
| Length | 109 |
| Max. P | 0.698808 |

| Location | 21,551,010 – 21,551,119 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 69.60 |
| Shannon entropy | 0.58597 |
| G+C content | 0.40356 |
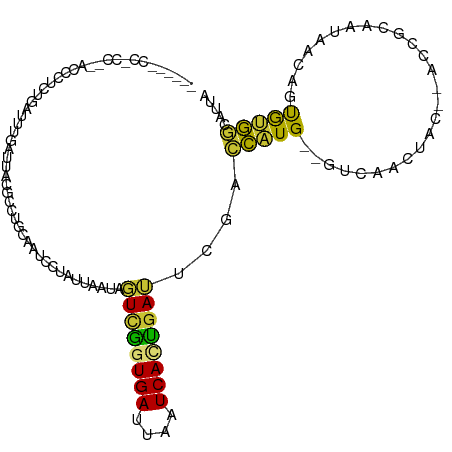
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -7.27 |
| Energy contribution | -6.80 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.698808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21551010 109 + 27905053 -CACCCCCGUCG-ACCAUCCGAUUUGAUUACAACUGCAAUCCUAUUAAUAGUCGGUGAUUAAUCACCGAUUCGACCAUG--GUCAACUAC--ACCGCAAUAACAGUAUGGCAUUA -.......((.(-(((((.(((...((((.(....).)))).........((((((((....)))))))))))...)))--))).))...--...((.(((....))).)).... ( -23.30, z-score = -1.63, R) >droSim1.chr3R 21386755 110 + 27517382 -UACCCCCACCACACCCUCUGAUUUGAUUACGCCUGCAAUCCUAUUAAUAGUCGGUGAUUAAUCACCGAUUCGACCAUG--GUCAACUAC--ACCGCAAUAACAGUGUGGCAUUA -........((((((..((......)).......(((............(((((((((....))))))))).(((....--)))......--...)))......))))))..... ( -26.90, z-score = -2.91, R) >droSec1.super_13 348021 110 + 2104621 -UACCCCCACCACACCCUCUGAUUUGAUUACGCCUCCAAUCCUAUUAAUAGUCGGUGAUUAAUCACCGAUUCGACCAUG--GUCAACUAC--ACCGCAAUAACUGUGUGGCAUUA -........((((((....((((..((((........))))..))))..(((((((((....))))))))).(((....--)))......--............))))))..... ( -25.50, z-score = -2.84, R) >droYak2.chr3R 3914131 109 - 28832112 CAAACUCCACCC--CUCUCUGAUUUGAUUACGCCUGCAAUCCUAUUAAUAGUCGGUGAUUAAUCACUGAUUCGACCAUG--GUCAACUAC--ACCGCAAUAACAGCAUGGCAUUA ............--.................((((((............(((((((((....))))))))).(((....--)))......--............))).))).... ( -22.80, z-score = -2.03, R) >droEre2.scaffold_4820 3982162 104 - 10470090 -----GCCCCUU--CCCCCUGAUCUUAUUACGCCUGCAAACCUAUUAAUAGUCAGUGAUUAAUCACUGAGUCGACCAUG--GUCAACUAC--ACCGCAAUAACAGCAUGGCAUUA -----.......--.................((((((..............(((((((....)))))))((.(((....--))).))...--............))).))).... ( -22.70, z-score = -2.58, R) >droAna3.scaffold_13340 12119506 97 - 23697760 -------------GCCCUCUGAUUUGAUUACGCGUGCAAUCCUAUUAAUAGU-AGUGAUUAAUCAUUGAUUGGCUUAUG--GUCAACUAC--ACGCUAAUAACAGUGGACUACCA -------------..((.(((.....((((.(((((.((((((((.....))-)).)))).........(((((.....--)))))...)--))))))))..))).))....... ( -21.60, z-score = -1.28, R) >dp4.chr2 7252511 96 - 30794189 -------------UCCCUUUGAUUCGAUUAC------AAUCUCCUCCAUAUUUGAUGAUUAAUCAAUGAAGUGUUUUUGUAGUUACCUAUUAACCAUAAUAGCCAUGGAAAAUUA -------------(((...((....((((((------((...(((.(((...((((.....))))))).)).)...))))))))..((((((....)))))).)).)))...... ( -14.20, z-score = -0.91, R) >droPer1.super_3 1062239 96 - 7375914 -------------UCCCUUUGAUUCGAUUAC------AAUCUCCUCCAUAUUUGAUGAUUAAUCAAUGAAGUGUUUUUGUAGUUACCUAUUAACCAUAAUAGCCAUGGAAAAUUA -------------(((...((....((((((------((...(((.(((...((((.....))))))).)).)...))))))))..((((((....)))))).)).)))...... ( -14.20, z-score = -0.91, R) >consensus ______CC_CC__ACCCUCUGAUUUGAUUACGCCUGCAAUCCUAUUAAUAGUCGGUGAUUAAUCACUGAUUCGACCAUG__GUCAACUAC__ACCGCAAUAACAGUGUGGCAUUA ..................................................((((((((....))))))))............................................. ( -7.27 = -6.80 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:54 2011