
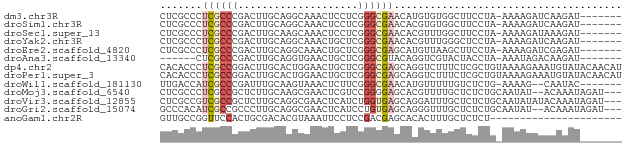
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,535,816 – 21,535,885 |
| Length | 69 |
| Max. P | 0.547152 |

| Location | 21,535,816 – 21,535,885 |
|---|---|
| Length | 69 |
| Sequences | 13 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 64.21 |
| Shannon entropy | 0.78157 |
| G+C content | 0.52801 |
| Mean single sequence MFE | -19.46 |
| Consensus MFE | -5.50 |
| Energy contribution | -5.76 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
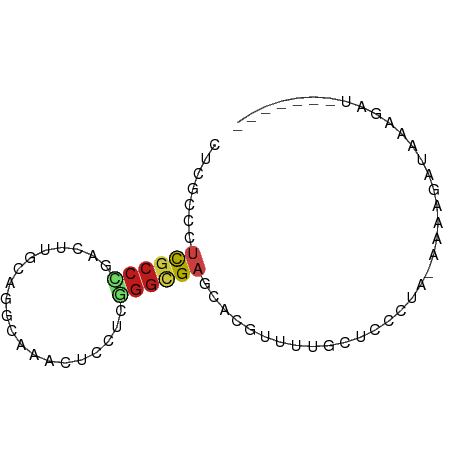
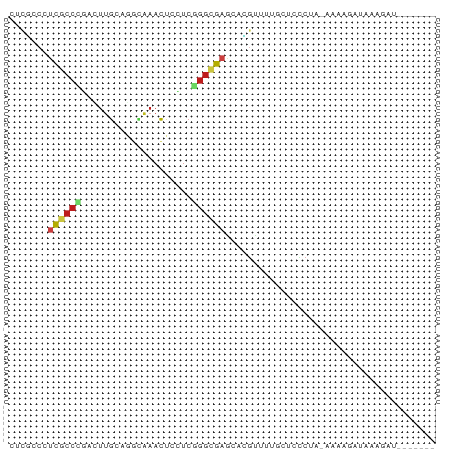
>dm3.chr3R 21535816 69 + 27905053 CUCGCCCUCGCCCGACUUGCAGGCAAACUCCUCGGGCGAACAUGUGUGGCUUCCUA-AAAAGAUCAAGAU------- ...(((.((((((((.(((....))).....))))))))((....)))))......-.............------- ( -17.70, z-score = -0.57, R) >droSim1.chr3R 21371637 69 + 27517382 CUCGCCCUCGCCCGACUUGCAGGCAAACUCCUCGGGCGAACACGUGUGGCUUCCUA-AAAAGAUCAAGAU------- ...(((.((((((((.(((....))).....))))))))((....)))))......-.............------- ( -17.70, z-score = -0.51, R) >droSec1.super_13 333076 69 + 2104621 CUCGCCCUCGCCCGACUUGCAAGCAAACUCCUCGGGCGAACACGUUUGGCUUCCUA-AAAAGAUAAAGAU------- ...(((.((((((((.(((....))).....))))))))........)))......-.............------- ( -16.10, z-score = -0.91, R) >droYak2.chr3R 3898481 69 - 28832112 CUCGCCCUCGCCCGACUUGCAGGCAAACUGCUCGGGCGAACACGUUUGGGCUCCUA-AAAAGAUCAAGAU------- ...((((((((((((...((((.....))))))))))))........)))).....-.............------- ( -26.60, z-score = -3.10, R) >droEre2.scaffold_4820 3967294 69 - 10470090 CUCGCCCUCGCCCGACUUGCAGGCAAACUGCUCGGGCGAGCAUGUUAAGCUUCCUA-AAAAGAUCGAGAU------- ((((..(((((((((...((((.....))))))))))))).........(((....-..)))..))))..------- ( -26.10, z-score = -2.81, R) >droAna3.scaffold_13340 12103994 63 - 23697760 ------CUCGCCCGACUUGCAGGUGAACUGCUCGGGCGUACAGGUCGUACUACCUA-AAAUAGACAAGAU------- ------..(((((((...((((.....)))))))))))...((((......)))).-.............------- ( -21.30, z-score = -2.51, R) >dp4.chr2 7234724 77 - 30794189 CACACCCUCGCCGGACUUGCACUGGAACUGCUCGGGCGAGCAGGUCUUUCUCGCUGUAAAAGAAAUGUAUACAACAU ...(((((((((.((...(((.......))))).))))))..))).......(.((((..(....)...)))).).. ( -17.50, z-score = 0.04, R) >droPer1.super_3 1044437 77 - 7375914 CACACCCUCGCCGGACUUGCACUGGAACUGCUCGGGCGAGCAGGUCUUUCUCGCUGUAAAAGAAAUGUAUACAACAU ...(((((((((.((...(((.......))))).))))))..))).......(.((((..(....)...)))).).. ( -17.50, z-score = 0.04, R) >droWil1.scaffold_181130 2153069 67 - 16660200 UUGACCAUCGCCCGAUUUGCAAGUAAACUCUUCGGGCGAACAUGUUUUUGUCUCUG-AAAAG--CAAUAC------- .......((((((((...(........)...))))))))...(((((((.......-)))))--))....------- ( -17.70, z-score = -2.44, R) >droMoj3.scaffold_6540 8494995 72 - 34148556 CUCGCCCUCGCCGCUCUUGCAAGCGAACUCGUCCGGGGAGCACGUUUUGCUCUCUGCAAUAU--ACAAAUAGAU--- .......((((.((....))..)))).......(((((((((.....)))))))))......--..........--- ( -18.50, z-score = -0.56, R) >droVir3.scaffold_12855 6961670 74 + 10161210 CUCGCCGUCGCCGCUCUUGCAGGCGAACUCAUCUGGUGAGCAGGAUUUGCUCUCUGCAAUAUAUACAAAUAGAU--- .(((((((((((((....)).))))).......))))))((((((.....)).)))).................--- ( -21.81, z-score = -1.27, R) >droGri2.scaffold_15074 1902196 72 - 7742996 GCCCACAUCGCCGCCCUUGCAGGCGAACUCAUCCUGUGAGCAGGGUUUGCUCUCUGCAAUAU--ACAAAUAGAU--- ((((...(((((((....)).))))).(((((...)))))..))))((((.....))))...--..........--- ( -21.10, z-score = -1.48, R) >anoGam1.chr2R 17280787 55 + 62725911 GUUGCCGGUUCCACUGCGACACGUAAAUUCCUCCGACGAGCACACUUUGCUCUCU---------------------- (((((.(......).)))))..............((.(((((.....))))))).---------------------- ( -13.40, z-score = -1.86, R) >consensus CUCGCCCUCGCCCGACUUGCAGGCAAACUCCUCGGGCGAGCACGUUUUGCUCCCUA_AAAAGAUAAAGAU_______ .......((((((....................))))))...................................... ( -5.50 = -5.76 + 0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:51 2011