
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,510,646 – 21,510,796 |
| Length | 150 |
| Max. P | 0.853869 |

| Location | 21,510,646 – 21,510,766 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.17 |
| Shannon entropy | 0.11745 |
| G+C content | 0.42667 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -19.36 |
| Energy contribution | -19.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21510646 120 + 27905053 UAGCCUGUACAUUGCACGCCACUCGCAUUCCUACCGCAGCGUAGAAAUUUAUAUUUUGGCUAAAAUUUAACAAAAUUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAA ..((.(((.....))).))...((((.......(((((((...(((((....))))).))).((((((....))))))....)))).............(((....)))))))....... ( -27.10, z-score = -1.96, R) >droSim1.chr3R 21340092 120 + 27517382 UAGCCUACACAUUGCACACCACUCGCAGUCCUACCACAGUGUAGAAAAUUAUAUUUUGGCUAAAAUUUAACAAAAUUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAA (((((.....(((((.........))))).((((......)))).............)))))((((((....))))))...(((((......)).....(((....))))))........ ( -24.80, z-score = -2.43, R) >droSec1.super_13 297422 120 + 2104621 UAGCCUACAUAUUGCACACCACUCGCAGUCCUACCGCAGCGCAGAAAAUUAUAUUUUGGCUAAAAUUUAACAAAAUUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAA ......................((((.......(((((((.(((((.......)))))))).((((((....))))))....)))).............(((....)))))))....... ( -26.00, z-score = -2.59, R) >droYak2.chr3R 3869866 120 - 28832112 UAGCCUUUACAGUGCACACCACUCGCAGUCCUACCGAAUCGCAGAAAAUAAUAUUUUGGCUAAAAUUUAACUAAAAUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAA (((((.....((((.....)))).((..((.....))...))...............)))))...................(((((......)).....(((....))))))........ ( -22.40, z-score = -1.32, R) >droEre2.scaffold_4820 3938843 120 - 10470090 UAGCCUUUACAGUGCACACCACUCGCAGUCCUACCGAAUCGCAGAAAAAUAUAUUUUGGCUAAAAUUUAACUAAAAUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAA (((((.....((((.....)))).((..((.....))...))...............)))))...................(((((......)).....(((....))))))........ ( -22.40, z-score = -1.32, R) >consensus UAGCCUAUACAUUGCACACCACUCGCAGUCCUACCGCAGCGCAGAAAAUUAUAUUUUGGCUAAAAUUUAACAAAAUUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAA (((((.......(((.........)))................((((......)))))))))...................(((((......)).....(((....))))))........ (-19.36 = -19.36 + -0.00)

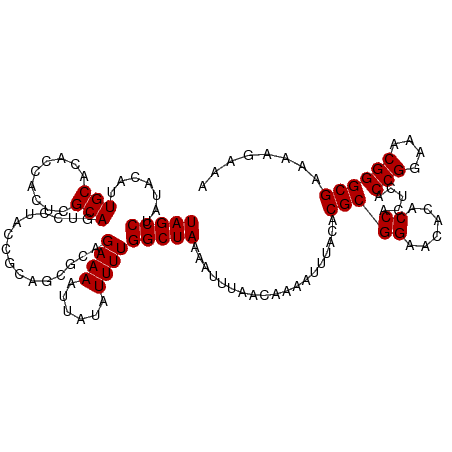

| Location | 21,510,676 – 21,510,796 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Shannon entropy | 0.08858 |
| G+C content | 0.40833 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -20.28 |
| Energy contribution | -19.72 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.853869 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 21510676 120 + 27905053 CUACCGCAGCGUAGAAAUUUAUAUUUUGGCUAAAAUUUAACAAAAUUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAAAGAAGGGGAGAUUUUAGAGAUGGCAAAACG ...(((((((...(((((....))))).))).((((((....))))))....))))......((((((.(((....)))............((((......)))).))).)))....... ( -23.90, z-score = -1.98, R) >droSim1.chr3R 21340122 120 + 27517382 CUACCACAGUGUAGAAAAUUAUAUUUUGGCUAAAAUUUAACAAAAUUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAAAGAAGGGGAGAUUUUAGAGAUGGCAAAACG ...(((.(((((((....))))))).)))(((((((((..(....(((.(.(((((......)).....(((....))))))....).)))....)..)))))))))............. ( -24.20, z-score = -2.61, R) >droSec1.super_13 297452 120 + 2104621 CUACCGCAGCGCAGAAAAUUAUAUUUUGGCUAAAAUUUAACAAAAUUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAAAGAAGGGGAGAUUUUAGAGAUGGCAAAACG ...(((((((.(((((.......)))))))).((((((....))))))....))))......((((((.(((....)))............((((......)))).))).)))....... ( -25.60, z-score = -2.74, R) >droYak2.chr3R 3869896 120 - 28832112 CUACCGAAUCGCAGAAAAUAAUAUUUUGGCUAAAAUUUAACUAAAAUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAAAGAAGGGGAGAUUUUAGAGAUGGCAAGACG ...(((..((((.((((......)))).))((((((((..((.........(((((......)).....(((....))))))............))..))))))))))..)))....... ( -21.70, z-score = -1.58, R) >droEre2.scaffold_4820 3938873 120 - 10470090 CUACCGAAUCGCAGAAAAAUAUAUUUUGGCUAAAAUUUAACUAAAAUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAAAGAAGGGGAGAUUUUGGAGAUGGCACAACG ...(((..((.(((((......(((((((.((.....)).)))))))....(((((......)).....(((....))))))...................)))))))..)))....... ( -21.30, z-score = -1.31, R) >consensus CUACCGCAGCGCAGAAAAUUAUAUUUUGGCUAAAAUUUAACAAAAUUUACACGCGGAACACACCACUCACCGGAAACGGGCGAAAAGAAAAGAAGGGGAGAUUUUAGAGAUGGCAAAACG ...(((.....(((((.......))))).(((((((((..(..........(((((......)).....(((....)))))).............)..)))))))))...)))....... (-20.28 = -19.72 + -0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:49 2011