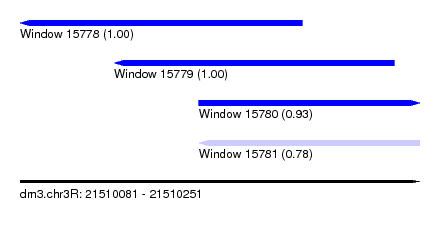
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,510,081 – 21,510,251 |
| Length | 170 |
| Max. P | 0.999411 |

| Location | 21,510,081 – 21,510,201 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.46 |
| Shannon entropy | 0.40136 |
| G+C content | 0.57778 |
| Mean single sequence MFE | -49.27 |
| Consensus MFE | -34.84 |
| Energy contribution | -34.31 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.53 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
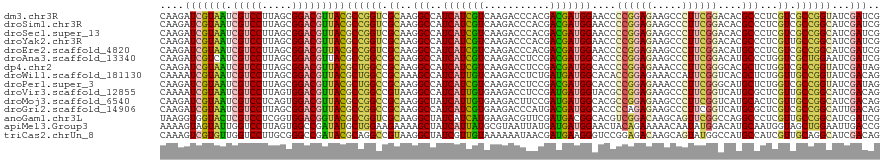
>dm3.chr3R 21510081 120 - 27905053 CAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGACGAUGGAACCCCGGAGAAGCCCUUCGGACACGCCCUCGUCGCCGGUAUCGAUCG ...((((((((.(((((.....)))))))))((((((.((.(((.(.((((((((.........))))))))....((((((.....))))))....).))).)).))))))...)))). ( -50.40, z-score = -3.67, R) >droSim1.chr3R 21339488 120 - 27517382 CAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGACGAUGGAACCCCGGAGAAGCCCUUCGGACACGCCCUCGUCGCCGGCAUCGAUCG ...((((((((.(((((.....)))))))))((((((.((.(((.(.((((((((.........))))))))....((((((.....))))))....).))).)).))))))...)))). ( -52.80, z-score = -4.35, R) >droSec1.super_13 296807 120 - 2104621 CAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGACGAUGGAACCCCGGAGAAGCCCUUCGGACACGCCCUCGUCGCCGGCAUCGAUCG ...((((((((.(((((.....)))))))))((((((.((.(((.(.((((((((.........))))))))....((((((.....))))))....).))).)).))))))...)))). ( -52.80, z-score = -4.35, R) >droYak2.chr3R 3869255 120 + 28832112 CAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGACGAUGGAACCCCGGAGAAGCCCUUCGGACACGCCCUCGUUGCCGGCAUCGAUCG ...((((((((.(((((.....)))))))))((((((.((.(((.(.((((((((.........))))))))....((((((.....))))))....).))).)).))))))...)))). ( -52.80, z-score = -4.51, R) >droEre2.scaffold_4820 3938237 120 + 10470090 CAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGACGAUGGAACCCCGGAGAAGCCCUUCGGACAUGCCCUCGUCGCCGGCAUCGAUCG ...((((((((.(((((.....)))))))))((((((.((.(((.((((((((((.........))))))))....((((((.....))))))...)).))).)).))))))...)))). ( -53.30, z-score = -4.56, R) >droAna3.scaffold_13340 12079694 120 + 23697760 CAAGAUCGUCAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGGCCAUCAUCGUCAAGACCUCCGACGAUGGCACCCCGGAGAAGCCCUUCGGACAUGCCCUGGUCGCUGGAAUCGAUCG ...(((((((..(((((.....))))).......((((....))))...........))((..((((.(.((((..((((((.....))))))...)))).).))))..))...))))). ( -48.40, z-score = -2.50, R) >dp4.chr2 7204915 120 + 30794189 CAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAGGCCAUCAUCGUCAAGACCUCCGACGAUGGCACCCCGGAGAAACCCUUCGGGCACGCUCUGGUCGCCGGUAUCGAUAG ...((((((((.(((((.....))))))))))..((((....)))))))((((.....(((..((((.(.(((..(((((((.....)))))))...))).).))))..)))..)))).. ( -51.20, z-score = -3.56, R) >droWil1.scaffold_181130 2122691 120 + 16660200 CAAAAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAAGCCAUCAUUGUCAAGACCUCUGAUGAUGGCACACCGGAGAAACCAUUCGGUCACGCUCUGGUUGCCGGUAUCGACAG .....((((((.(((((.....)))))))))((((((......(((((((((............)))))))))..(((((((..(((....)))....))))))).))))))...))... ( -45.30, z-score = -3.60, R) >droPer1.super_3 1011890 120 + 7375914 CAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAGGCCAUCAUCGUCAAGACCUCCGACGAUGGCACCCCGGAGAAACCCUUCGGGCAUGCUCUGGUCGCCGGUAUCGAUAG ...((((((((.(((((.....))))))))))..((((....)))))))((((.....(((..((((.(.((((.(((((((.....)))))))..)))).).))))..)))..)))).. ( -52.60, z-score = -3.94, R) >droVir3.scaffold_12855 6923115 120 - 10161210 CAAAAUCGUAAUCGUCCUUAGUGGACGUUACGCCGGCCGUAAGGCCAUCAUUGUGAAGACCUCCGAUGAUGGUACGCCGGAGAAGCCCUUCGGUCAUGCGCUCGUUGCCGGCAUCGACAG .....((((((.(((((.....)))))))))((((((.(..((((((((((((.((.....))))))))))))..(((((((.....)))))))......))..).))))))...))... ( -51.00, z-score = -3.92, R) >droMoj3.scaffold_6540 8459071 120 + 34148556 CAAGAUCGUAAUCGUCCUCAGUGGACGUUACGCCGGCCGCAAGGCUAUCAUUGUGAAGACUUCCGAUGAUGGCACGCCGGAGAAGCCCUUCGGUCAUGCACUCGUUGCCGGCAUCGACAG ...(((.((((.(((((.....)))))))))((((((.((.((((((((((((.((.....))))))))))))..(((((((.....)))))))......)).)).)))))))))..... ( -52.70, z-score = -4.05, R) >droGri2.scaffold_14906 3063409 120 - 14172833 CAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGGCCAUCAUCGUGAAGACCCAUGACGAUGGCACCCCAGAGAAGCCCUUCGGUCAUGCGCUCGUCGCCGGCAUUGACAG (((....((((.(((((.....)))))))))((((((.((...((((((.(((((......))))).))))))......(((..((.(....)....)).))))).)))))).))).... ( -47.90, z-score = -3.32, R) >anoGam1.chr3L 30714694 120 - 41284009 UAAGGUGGUACUCGUCCUCGGUGGACGGUACGCCGGUCGCAAGGCUAUCAUCAUGAAGACGUUCGAUGACGGCACGUCGGACAAGCAGUUCGGCCAGGCCCUCGUUGCCGGCAUCGAUCG ...((((((((.(((((.....)))))))))...((((....))))..)))).......((.((((((.(((((((..((....((......))....))..)).))))).)))))).)) ( -49.00, z-score = -1.59, R) >apiMel3.Group3 8878051 120 + 12341916 AAAAGUAGUAUUGGUCCUUAGUGGCCGAUAUGCUGGAAGAAAAGCUAUCAUUAUGCGUAAUUAUGAUGAUGGAACUACAGAAAAACAAUAUGGACAUGCAAUGGUAGCUGGAAUUGACCG ...((((.((((((((......))))))))))))((..((..(((((((((..(((((..(((((.((.((......))......)).)))))..))))))))))))))....))..)). ( -28.60, z-score = -1.78, R) >triCas2.chrUn_8 595500 120 + 609019 CAAAGUCGUGUUGGUCCUUGCGGGCCGAUACGCAGGCCGUAAGGCUAUCGUUGUAAAAAAUAACGAUGAAGGGUCCGGAGACAAGCAGUAUGGCCAUGCCAUCGUUGCAGGCAUCGACAG ....(((((((((((((....))))))))))...(((((((..(((((((((((.....)))))))).........(....).)))..)))))))(((((.........))))).))).. ( -50.20, z-score = -4.34, R) >consensus CAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGGCCAUCAUCGUCAAGACCUACGACGAUGGCACCCCGGAGAAGCCCUUCGGACAUGCCCUCGUCGCCGGCAUCGAUCG ....(((((((.(((((.....)))))))))((((((.((..(((..(((((((...........)))))))....((((((.....))))))....)))...)).))))))...))).. (-34.84 = -34.31 + -0.53)
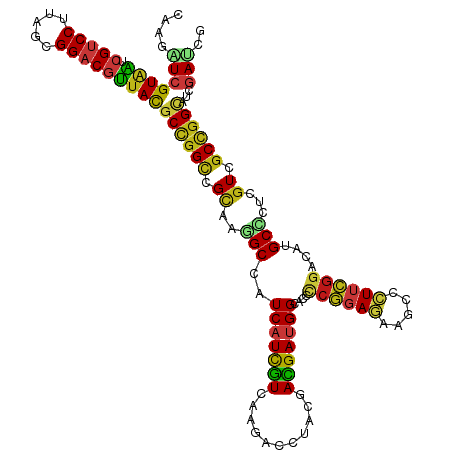
| Location | 21,510,121 – 21,510,240 |
|---|---|
| Length | 119 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.90 |
| Shannon entropy | 0.47170 |
| G+C content | 0.52928 |
| Mean single sequence MFE | -39.71 |
| Consensus MFE | -27.26 |
| Energy contribution | -26.16 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.996517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
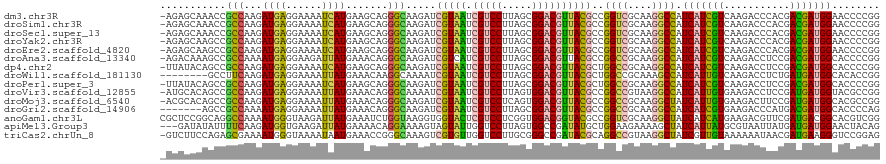
>dm3.chr3R 21510121 119 - 27905053 -AGAGCAAACCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGACGAUGGAACCCCGG -........((((....((((......))))...)).(((......(((((.(((((.....))))))))))..((((....)))).((((((((.........))))))))...))))) ( -39.20, z-score = -2.10, R) >droSim1.chr3R 21339528 119 - 27517382 -AGAGCAAACCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGACGAUGGAACCCCGG -........((((....((((......))))...)).(((......(((((.(((((.....))))))))))..((((....)))).((((((((.........))))))))...))))) ( -39.20, z-score = -2.10, R) >droSec1.super_13 296847 119 - 2104621 -AGAGCAAACCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGACGAUGGAACCCCGG -........((((....((((......))))...)).(((......(((((.(((((.....))))))))))..((((....)))).((((((((.........))))))))...))))) ( -39.20, z-score = -2.10, R) >droYak2.chr3R 3869295 119 + 28832112 -AGAGCAAGCCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGACGAUGGAACCCCGG -.(.((..(((((....((((......))))...))..)))......((((.(((((.....))))))))))))((((....)))).((((((((.........))))))))........ ( -38.90, z-score = -1.75, R) >droEre2.scaffold_4820 3938277 119 + 10470090 -AGAGCAAGCCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGACGAUGGAACCCCGG -.(.((..(((((....((((......))))...))..)))......((((.(((((.....))))))))))))((((....)))).((((((((.........))))))))........ ( -38.90, z-score = -1.75, R) >droAna3.scaffold_13340 12079734 119 + 23697760 -AGACAAAGCCGCCAAAAUGAGGAAGAUUAUGAAACAGGGCAAGAUCGUCAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGGCCAUCAUCGUCAAGACCUCCGACGAUGGCACCCCGG -.(((....((((.....((((((.(((.((((............)))).))).))))))))))..)))..(((((((....))))...((((((.........)))))))))....... ( -39.80, z-score = -1.98, R) >dp4.chr2 7204955 119 + 30794189 -UUAUACAGCCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAGGCCAUCAUCGUCAAGACCUCCGACGAUGGCACCCCGG -........((((....((((......))))...)).(((.......((((.(((((.....)))))))))(((((((....))))).(((((((.........)))))))))..))))) ( -42.70, z-score = -2.71, R) >droWil1.scaffold_181130 2122731 112 + 16660200 --------GCCUUCAAGAUGAGGAAAAUUAUGAAACAAGGCAAAAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAAGCCAUCAUUGUCAAGACCUCUGAUGAUGGCACACCGG --------((((((.....))))...............(((.....(((((.(((((.....))))))))))...)))))...(((((((((............)))))))))....... ( -34.70, z-score = -2.56, R) >droPer1.super_3 1011930 119 + 7375914 -UUAUACAGCCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAGGCCAUCAUCGUCAAGACCUCCGACGAUGGCACCCCGG -........((((....((((......))))...)).(((.......((((.(((((.....)))))))))(((((((....))))).(((((((.........)))))))))..))))) ( -42.70, z-score = -2.71, R) >droVir3.scaffold_12855 6923155 119 - 10161210 -AUGCACAGCCGCCAAGAUGAGGAAAAUUAUGAAACAGGGCAAAAUCGUAAUCGUCCUUAGUGGACGUUACGCCGGCCGUAAGGCCAUCAUUGUGAAGACCUCCGAUGAUGGUACGCCGG -.........(((((...((((((..(((((((............)))))))..)))))).))).)).....((((((....)((((((((((.((.....))))))))))))..))))) ( -41.50, z-score = -2.69, R) >droMoj3.scaffold_6540 8459111 119 + 34148556 -ACGCACAGCCGCCAAGAUGAGGAAAAUUAUGAAACAGGGCAAGAUCGUAAUCGUCCUCAGUGGACGUUACGCCGGCCGCAAGGCUAUCAUUGUGAAGACUUCCGAUGAUGGCACGCCGG -.........(((((...((((((..(((((((............)))))))..)))))).))).)).....((((((....)((((((((((.((.....))))))))))))..))))) ( -44.20, z-score = -2.92, R) >droGri2.scaffold_14906 3063449 113 - 14172833 -------AGCCGCCAAAAUGAGGAAAAUUAUGAAACAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGGCCAUCAUCGUGAAGACCCAUGACGAUGGCACCCCAG -------.(((((.....((((((..(((((((............)))))))..)))))))))).).....(((((((....))))(((.(((((......))))).))))))....... ( -40.90, z-score = -3.58, R) >anoGam1.chr3L 30714734 120 - 41284009 CGCUCCGGCAGGCCAAAAUGGGUAAGAUUAUGAAAUCUGGUAAGGUGGUACUCGUCCUCGGUGGACGGUACGCCGGUCGCAAGGCUAUCAUCAUGAAGACGUUCGAUGACGGCACGUCGG ....(((((.((((..........(((((....))))).....((((.(((.(((((.....)))))))))))))))).....((..(((((..(((....))))))))..))..))))) ( -42.00, z-score = -1.10, R) >apiMel3.Group3 8878091 117 + 12341916 ---GAUAUAUUUUCAAGAUGGUGAAGAUUAUGAAAACAGGAAAAGUAGUAUUGGUCCUUAGUGGCCGAUAUGCUGGAAGAAAAGCUAUCAUUAUGCGUAAUUAUGAUGAUGGAACUACAG ---..((((((((((......)))))).))))............((((((((((((......)))))))...............(((((((((((......))))))))))).))))).. ( -26.30, z-score = -2.02, R) >triCas2.chrUn_8 595540 119 + 609019 -GUCUUCCAGAGCGAAAAUGGGUAAAAUAAUGAAACCGGGCAAAGUCGUGUUGGUCCUUGCGGGCCGAUACGCAGGCCGUAAGGCUAUCGUUGUAAAAAAUAACGAUGAAGGGUCCGGAG -....((((.........)))).............((((((...(.(((((((((((....)))))))))))).((((....))))((((((((.....)))))))).....)))))).. ( -45.40, z-score = -4.06, R) >consensus _AGAGCAAGCCGCCAAGAUGAGGAAAAUCAUGAAACAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGGCCAUCAUCGUCAAGACCUACGACGAUGGCACCCCGG ...........(((...((((......)))).......))).....(((((.(((((.....))))))))))..((((....)))).(((((((...........)))))))........ (-27.26 = -26.16 + -1.10)
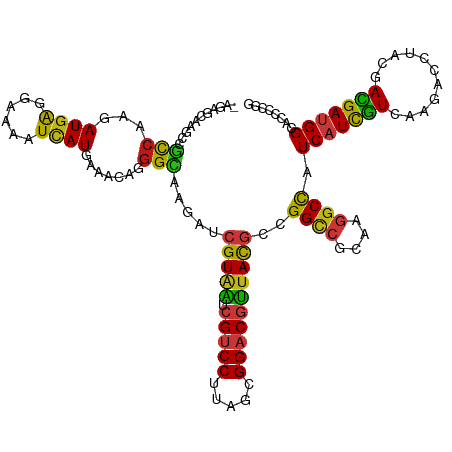
| Location | 21,510,157 – 21,510,251 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Shannon entropy | 0.53554 |
| G+C content | 0.51131 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -15.77 |
| Energy contribution | -15.45 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
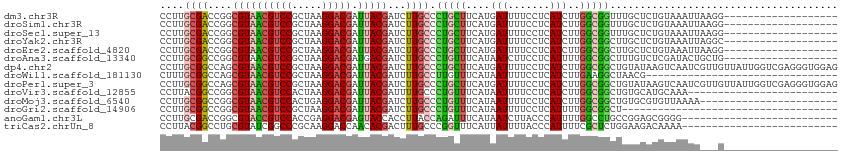
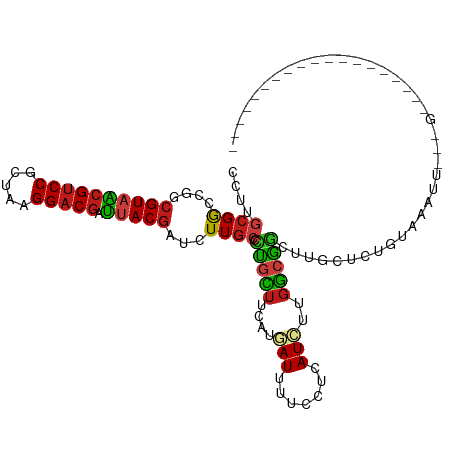
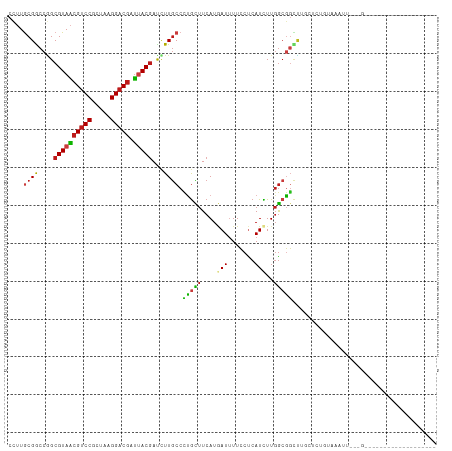
>dm3.chr3R 21510157 94 + 27905053 CCUUGCGACCGGCGUAACGUCCGCUAAGGACGAUUACGAUCUUGCCCUGCUUCAUGAUUUUCCUCAUCUUGGCGGUUUGCUCUGUAAAUUAAGG------------------- ((((((((....((((((((((.....))))).)))))...)))).(((((..((((......))))...)))))...............))))------------------- ( -26.20, z-score = -1.61, R) >droSim1.chr3R 21339564 94 + 27517382 CCUUGCGACCGGCGUAACGUCCGCUAAGGACGAUUACGAUCUUGCCCUGCUUCAUGAUUUUCCUCAUCUUGGCGGUUUGCUCUGUAAAUUAAGG------------------- ((((((((....((((((((((.....))))).)))))...)))).(((((..((((......))))...)))))...............))))------------------- ( -26.20, z-score = -1.61, R) >droSec1.super_13 296883 94 + 2104621 CCUUGCGACCGGCGUAACGUCCGCUAAGGACGAUUACGAUCUUGCCCUGCUUCAUGAUUUUCCUCAUCUUGGCGGUUUGCUCUGUAAAUUAAGG------------------- ((((((((....((((((((((.....))))).)))))...)))).(((((..((((......))))...)))))...............))))------------------- ( -26.20, z-score = -1.61, R) >droYak2.chr3R 3869331 94 - 28832112 CCUUGCGACCGGCGUAACGUCCGCUAAGGACGAUUACGAUCUUGCCCUGCUUCAUGAUUUUCCUCAUCUUGGCGGCUUGCUCUGUAAAUUAGGC------------------- ....((((....((((((((((.....))))).)))))...)))).(((((..((((......))))...)))))...((.(((.....)))))------------------- ( -25.40, z-score = -0.81, R) >droEre2.scaffold_4820 3938313 94 - 10470090 CCUUGCGACCGGCGUAACGUCCGCUAAGGACGAUUACGAUCUUGCCCUGCUUCAUGAUUUUCCUCAUCUUGGCGGCUUGCUCUGUAAAUUAAGG------------------- ((((((((....((((((((((.....))))).)))))...)))).(((((..((((......))))...)))))...............))))------------------- ( -26.00, z-score = -1.46, R) >droAna3.scaffold_13340 12079770 94 - 23697760 CCUUGCGGCCGGCGUAACGUCCGCUAAGGACGAUGACGAUCUUGCCCUGUUUCAUAAUCUUCCUCAUUUUGGCGGCUUUGUCUCGAUACUGCUG------------------- ((((((((.((......)).)))).)))).(((.(((((..(((((........................)))))..)))))))).........------------------- ( -26.36, z-score = -1.29, R) >dp4.chr2 7204991 113 - 30794189 CCUUGCGGCCAGCGUAACGUCCGCUAAGGACGAUUACGAUCUUGCCCUGCUUCAUGAUUUUCCUCAUCUUGGCGGCUGUAUAAGUCAAUCGUUGUUAUUGGUCGAGGGUGGAG ((((.(((((((.(((((((((.....))))).))))(((..(((.(((((..((((......))))...)))))..)))...)))...........)))))))))))..... ( -35.60, z-score = -1.10, R) >droWil1.scaffold_181130 2122767 81 - 16660200 CUUUGCGGCCAGCGUAACGUCCGCUAAGGACGAUUACGAUUUUGCCUUGUUUCAUAAUUUUCCUCAUCUUGAAGGCUAACG-------------------------------- ....(((((...((((((((((.....))))).))))).....)))...(((((...............))))))).....-------------------------------- ( -20.46, z-score = -1.75, R) >droPer1.super_3 1011966 113 - 7375914 CCUUGCGGCCAGCGUAACGUCCGCUAAGGACGAUUACGAUCUUGCCCUGCUUCAUGAUUUUCCUCAUCUUGGCGGCUGUAUAAGUCAAUCGUUGUUAUUGGUCGAGGGUGGAG ((((.(((((((.(((((((((.....))))).))))(((..(((.(((((..((((......))))...)))))..)))...)))...........)))))))))))..... ( -35.60, z-score = -1.10, R) >droVir3.scaffold_12855 6923191 88 + 10161210 CCUUACGGCCGGCGUAACGUCCACUAAGGACGAUUACGAUUUUGCCCUGUUUCAUAAUUUUCCUCAUCUUGGCGGCUGUGCAUGCAAA------------------------- ...(((((((..((((((((((.....))))).))))).....(((..(..................)..))))))))))........------------------------- ( -25.97, z-score = -2.10, R) >droMoj3.scaffold_6540 8459147 90 - 34148556 CCUUGCGGCCGGCGUAACGUCCACUGAGGACGAUUACGAUCUUGCCCUGUUUCAUAAUUUUCCUCAUCUUGGCGGCUGUGCGUGUUAAAA----------------------- ....((((((((((((((((((.....))))).))))).))..(((..(..................)..)))))))))...........----------------------- ( -25.87, z-score = -1.14, R) >droGri2.scaffold_14906 3063485 77 + 14172833 CCUUGCGGCCGGCGUAACGUCCGCUAAGGACGAUUACGAUCUUGCCCUGUUUCAUAAUUUUCCUCAUUUUGGCGGCU------------------------------------ ......((((((((((((((((.....))))).))))).))..(((........................)))))))------------------------------------ ( -21.56, z-score = -0.96, R) >anoGam1.chr3L 30714770 87 + 41284009 CCUUGCGACCGGCGUACCGUCCACCGAGGACGAGUACCACCUUACCAGAUUUCAUAAUCUUACCCAUUUUGGCCUGCCGGAGCGGGG-------------------------- ((((((..((((((((((((((.....))))).)))).......(((((..................)))))...))))).))))))-------------------------- ( -33.67, z-score = -3.67, R) >triCas2.chrUn_8 595576 87 - 609019 CCUUACGGCCUGCGUAUCGGCCCGCAAGGACCAACACGACUUUGCCCGGUUUCAUUAUUUUACCCAUUUUCGCUCUGGAAGACAAAA-------------------------- ......(((..((((...((.((....)).))...))).)...))).(((...........)))..(((((......))))).....-------------------------- ( -16.00, z-score = 0.74, R) >consensus CCUUGCGGCCGGCGUAACGUCCGCUAAGGACGAUUACGAUCUUGCCCUGCUUCAUGAUUUUCCUCAUCUUGGCGGCUUGCUCUGUAAAUU___G___________________ ....((((....((((((((((.....))))).)))))...)))).(((((....(((.......)))..)))))...................................... (-15.77 = -15.45 + -0.32)
| Location | 21,510,157 – 21,510,251 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.96 |
| Shannon entropy | 0.53554 |
| G+C content | 0.51131 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -17.93 |
| Energy contribution | -17.84 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.776934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
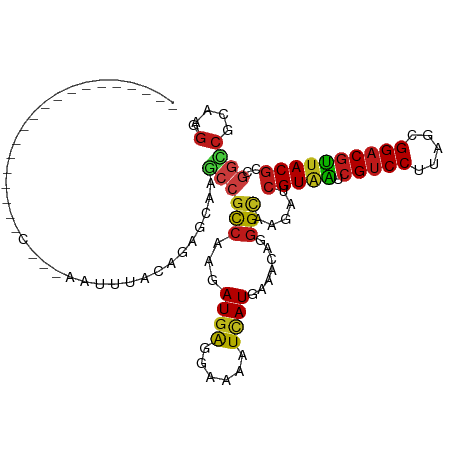
>dm3.chr3R 21510157 94 - 27905053 -------------------CCUUAAUUUACAGAGCAAACCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGG -------------------..............((..((((((...((((......)))).......))).....(((((.(((((.....))))))))))..))).)).... ( -22.80, z-score = -0.47, R) >droSim1.chr3R 21339564 94 - 27517382 -------------------CCUUAAUUUACAGAGCAAACCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGG -------------------..............((..((((((...((((......)))).......))).....(((((.(((((.....))))))))))..))).)).... ( -22.80, z-score = -0.47, R) >droSec1.super_13 296883 94 - 2104621 -------------------CCUUAAUUUACAGAGCAAACCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGG -------------------..............((..((((((...((((......)))).......))).....(((((.(((((.....))))))))))..))).)).... ( -22.80, z-score = -0.47, R) >droYak2.chr3R 3869331 94 + 28832112 -------------------GCCUAAUUUACAGAGCAAGCCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGG -------------------((((.......)).))..((.(((...((((......)))).......(((......((((.(((((.....))))))))))))))).)).... ( -23.30, z-score = 0.02, R) >droEre2.scaffold_4820 3938313 94 + 10470090 -------------------CCUUAAUUUACAGAGCAAGCCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGG -------------------..............((..((((((...((((......)))).......))).....(((((.(((((.....))))))))))..))).)).... ( -23.10, z-score = -0.24, R) >droAna3.scaffold_13340 12079770 94 + 23697760 -------------------CAGCAGUAUCGAGACAAAGCCGCCAAAAUGAGGAAGAUUAUGAAACAGGGCAAGAUCGUCAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGG -------------------..((.((..((.(((....((((.....((((((.(((.((((............)))).))).))))))))))..))).))...)).)).... ( -22.60, z-score = -0.06, R) >dp4.chr2 7204991 113 + 30794189 CUCCACCCUCGACCAAUAACAACGAUUGACUUAUACAGCCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAGG .............((((.......)))).......((((.(((...((((......)))).......)))......((((.(((((.....))))))))))))).((....)) ( -26.00, z-score = 0.04, R) >droWil1.scaffold_181130 2122767 81 + 16660200 --------------------------------CGUUAGCCUUCAAGAUGAGGAAAAUUAUGAAACAAGGCAAAAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAAG --------------------------------.....((((((.....))))...............(((.....(((((.(((((.....))))))))))...))))).... ( -22.20, z-score = -1.96, R) >droPer1.super_3 1011966 113 + 7375914 CUCCACCCUCGACCAAUAACAACGAUUGACUUAUACAGCCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAGG .............((((.......)))).......((((.(((...((((......)))).......)))......((((.(((((.....))))))))))))).((....)) ( -26.00, z-score = 0.04, R) >droVir3.scaffold_12855 6923191 88 - 10161210 -------------------------UUUGCAUGCACAGCCGCCAAGAUGAGGAAAAUUAUGAAACAGGGCAAAAUCGUAAUCGUCCUUAGUGGACGUUACGCCGGCCGUAAGG -------------------------.........((.((((((...((((......)))).......))).....(((((.(((((.....))))))))))..))).)).... ( -22.50, z-score = -0.49, R) >droMoj3.scaffold_6540 8459147 90 + 34148556 -----------------------UUUUAACACGCACAGCCGCCAAGAUGAGGAAAAUUAUGAAACAGGGCAAGAUCGUAAUCGUCCUCAGUGGACGUUACGCCGGCCGCAAGG -----------------------.........((..(((..(((...((((((..(((((((............)))))))..)))))).)))..)))..))...((....)) ( -24.40, z-score = -1.15, R) >droGri2.scaffold_14906 3063485 77 - 14172833 ------------------------------------AGCCGCCAAAAUGAGGAAAAUUAUGAAACAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGG ------------------------------------.((.(((....((((((..(((((((............)))))))..))))))(((.......))).))).)).... ( -24.00, z-score = -1.71, R) >anoGam1.chr3L 30714770 87 - 41284009 --------------------------CCCCGCUCCGGCAGGCCAAAAUGGGUAAGAUUAUGAAAUCUGGUAAGGUGGUACUCGUCCUCGGUGGACGGUACGCCGGUCGCAAGG --------------------------....((.(((((..(((......))).(((((....))))).........((((.(((((.....))))))))))))))..)).... ( -31.00, z-score = -0.82, R) >triCas2.chrUn_8 595576 87 + 609019 --------------------------UUUUGUCUUCCAGAGCGAAAAUGGGUAAAAUAAUGAAACCGGGCAAAGUCGUGUUGGUCCUUGCGGGCCGAUACGCAGGCCGUAAGG --------------------------((((((........))))))...(((...........))).(((...(.(((((((((((....))))))))))))..)))...... ( -25.90, z-score = -0.81, R) >consensus ___________________C___AAUUUACAGAGCAAGCCGCCAAGAUGAGGAAAAUCAUGAAACAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGG .....................................((((((...((((......)))).......))).....(((((.(((((.....))))))))))..)))(....). (-17.93 = -17.84 + -0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:48 2011