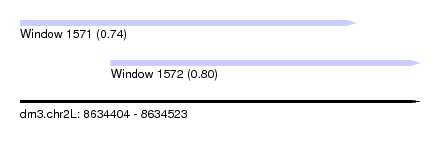
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,634,404 – 8,634,523 |
| Length | 119 |
| Max. P | 0.803635 |

| Location | 8,634,404 – 8,634,504 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.62 |
| Shannon entropy | 0.70146 |
| G+C content | 0.46827 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -9.39 |
| Energy contribution | -9.38 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
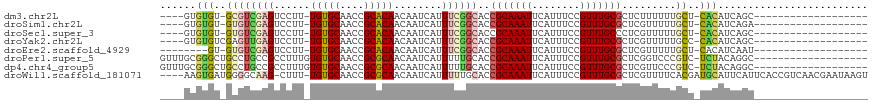
>dm3.chr2L 8634404 100 + 23011544 ------------------UGUUCUUAUCCUGUAGCCUGCAUUUGUGUGUGUGCGU--CGAGUCCUUUGUGCAACCGCACAACAAUCAUUUCGGCACCGCAAAUUCAUUUCCGUUUGCGCU ------------------...............((.(((((....))))).))((--((((....((((((....)))))).......))))))..(((((((........))))))).. ( -24.60, z-score = -1.01, R) >droSim1.chr2L 8416987 100 + 22036055 ------------------UGUUCUUAUCCUGUAUCCUGCAUUUGUGUGUGUGUGU--CGAGUCCUUUGUGCAACCGCACAACAAUCAUUUCGGCACCGCAAAUUCAUUUCCGUUUGCGCU ------------------...................((((......))))((((--((((....((((((....)))))).......))))))))(((((((........))))))).. ( -24.40, z-score = -1.80, R) >droSec1.super_3 4113857 102 + 7220098 ------------------UGUUCUUAUCCUGUAUCCUGCAUUUGUGUGUGUGUGUGUCGAGUCCUUUGUGCAACCGCACAACAAUCAUUUCGGCACCGCAAAUUCAUUUCCGUUUGCCCU ------------------...................(((..((.(((((((.((((((((....((((((....)))))).......))))))))))))....)))...))..)))... ( -24.80, z-score = -2.37, R) >droYak2.chr2L 11277175 96 + 22324452 ------------------UGUUCUUAUCCUG-----CGCAUUUGUGUGUGU-CGAGUUGAGUCCUUUGUGCAACCGCACAACAAUCAUUUCGGCACCGCAAAUUCAUUUCCGUUUGCGCU ------------------............(-----((((((((((.((((-((((.(((.....((((((....))))))...))).))))))))))))))............))))). ( -33.20, z-score = -4.22, R) >droEre2.scaffold_4929 9226466 91 + 26641161 ------------------UGUUCUUAUCCUG-----UGCAUUUGUGUGUGU------CGAGUCCUUUGUGCAACCGCACAACAAUCAUUUCGGCACCGCAAAUUCAUUUCCGUUUGCGCU ------------------............(-----((((((((((.((((------((((....((((((....)))))).......))))))))))))))............))))). ( -27.30, z-score = -3.31, R) >droAna3.scaffold_12916 10274882 90 + 16180835 ------------------CGUCCUUGUUCCUUG---UGCAUGUGUGUGUGUGUGA---GAG-CCUUUGUGCUACCGCAUAACAAUCUCUUUUGCACAGCGAUUUCAUUUCGGUUC----- ------------------((....((.((.(((---((((.(..(((.((((((.---.((-(......)))..)))))))))....)...))))))).))...))...))....----- ( -21.60, z-score = -0.94, R) >droPer1.super_5 1368122 118 - 6813705 CGUUCUUGUUCUUCUCCUCUGCGUCAGGCAGGAGGAUCUGUUUGCGGGCUGCCUG--CCGCCUUUGUGUGCAACCGCGCAACAAUCAUUUUUGCACCGCAAAUUCAUUUCCGUUUGCGCU ....................(((.(((((.(((((....((((((((((.((...--..))..((((((((....)))).))))........)).))))))))...))))))))))))). ( -34.70, z-score = -0.42, R) >dp4.chr4_group5 1397454 118 - 2436548 CGUCCUUGUUCUUCUCCUCUGCGUCAGGCAGGAGGAUCUGUUUGCGGGCUGCCUG--CCGCCUUUGUGUGCAACCGCGCAACAAUCAUUUUUGCACCGCAAAUUCAUUUCCGUUUGCGCU ....................(((.(((((.(((((....((((((((((.((...--..))..((((((((....)))).))))........)).))))))))...))))))))))))). ( -34.70, z-score = -0.39, R) >droWil1.scaffold_181071 1054870 90 + 1211509 ---------------------------UUCUUGUGGUAUGUU-GAAAGUGAUGGG--GCAAGCUUUUGUGCAACCGCGCAACAAUCAUUUUUGCACCGCAAAUUCAUUUCCGUUUGCGCU ---------------------------...(((((((..((.-(((((((((.((--(....)))((((((....))))))..))))))))))))))))))................... ( -20.30, z-score = 1.02, R) >droVir3.scaffold_12963 14485307 98 - 20206255 ---------------------UGUUG-CGCUGCGUCCAUCUUUUAUUUCAUUUCGUGCUUUGCCUUUGUGCAACUGUGCAACAAUCAUUUUUGCACCGCAAAUUCAUUUCCGUUUGCACU ---------------------.((((-(((.(((..(((...............)))...)))....))))))).((((((.........)))))).((((((........))))))... ( -20.76, z-score = -1.35, R) >droGri2.scaffold_15252 7730415 99 + 17193109 ---------------------CGUCGUCGUUGCUUCAAUCUUUUAUUUCAUUCCAUUCUUUGCCGUUGUGCAACUGUGCAACAAUCAGUUUUGCACCGAAAAUUCAUUUCCGCUUGCACU ---------------------.......(((((..((((.........................)))).))))).(((((((((......)))....((((.....))))...)))))). ( -12.91, z-score = 0.47, R) >consensus __________________UGUUCUUAUCCUGUAG_CUGCAUUUGUGUGUGUGCGG__CGAGUCCUUUGUGCAACCGCACAACAAUCAUUUUUGCACCGCAAAUUCAUUUCCGUUUGCGCU .................................................................((((((....))))))................((((((........))))))... ( -9.39 = -9.38 + -0.01)



| Location | 8,634,431 – 8,634,523 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.92 |
| Shannon entropy | 0.41974 |
| G+C content | 0.49697 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.74 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
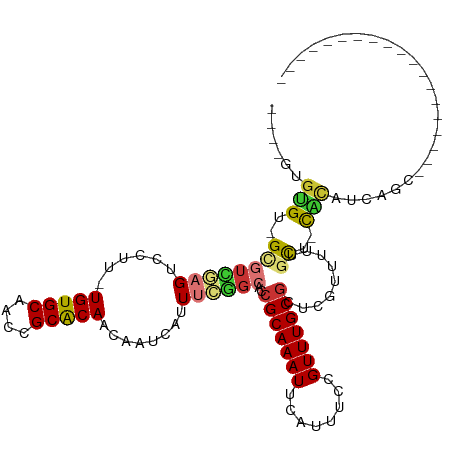
>dm3.chr2L 8634431 92 + 23011544 ----GUGUGU-GCGUCGAGUCCUU-UGUGCAACCGCACAACAAUCAUUUCGGCACCGCAAAUUCAUUUCCGUUUGCGCUCUUUUUUGCU-CACAUCAGC------------------- ----(((((.-((((((((....(-(((((....)))))).......))))))..(((((((........))))))).........)).-)))))....------------------- ( -27.70, z-score = -3.15, R) >droSim1.chr2L 8417014 92 + 22036055 ----GUGUGU-GUGUCGAGUCCUU-UGUGCAACCGCACAACAAUCAUUUCGGCACCGCAAAUUCAUUUCCGUUUGCGCUCGUUUUUGCU-CACAUCAGA------------------- ----(((.((-((((((((....(-(((((....)))))).......))))))))(((((((........))))))).........)).-)))......------------------- ( -27.20, z-score = -2.51, R) >droSec1.super_3 4113886 92 + 7220098 ----GUGUGU-GUGUCGAGUCCUU-UGUGCAACCGCACAACAAUCAUUUCGGCACCGCAAAUUCAUUUCCGUUUGCCCUCGUUUUUGCU-CACAUCAGC------------------- ----(((.((-((((((((....(-(((((....)))))).......)))))))).((((((........))))))..........)).-)))......------------------- ( -25.20, z-score = -2.69, R) >droYak2.chr2L 11277197 93 + 22324452 ----GUGUGUCGAGUUGAGUCCUU-UGUGCAACCGCACAACAAUCAUUUCGGCACCGCAAAUUCAUUUCCGUUUGCGCUCGUUUUUGCC-CACAUCAGC------------------- ----(.((((((((.(((.....(-(((((....))))))...))).)))))))))((((((........))))))((........)).-.........------------------- ( -26.80, z-score = -2.59, R) >droEre2.scaffold_4929 9226488 88 + 26641161 --------GU-GUGUCGAGUCCUU-UGUGCAACCGCACAACAAUCAUUUCGGCACCGCAAAUUCAUUUCCGUUUGCGCUCGUUUUUGCU-CACAUCAAU------------------- --------(.-((((((((....(-(((((....)))))).......)))))))))((((((........))))))((........)).-.........------------------- ( -23.20, z-score = -2.73, R) >droPer1.super_5 1368161 98 - 6813705 GUUUGCGGGCUGCCUGCCGCCUUUGUGUGCAACCGCGCAACAAUCAUUUUUGCACCGCAAAUUCAUUUCCGUUUGCGCUCGGUCCCGUC-UCUACAGGC------------------- ((..(((((..(((((((((....))).)))...((((((........(((((...)))))...........))))))..)))))))).-...))....------------------- ( -25.51, z-score = 0.42, R) >dp4.chr4_group5 1397493 98 - 2436548 GUUUGCGGGCUGCCUGCCGCCUUUGUGUGCAACCGCGCAACAAUCAUUUUUGCACCGCAAAUUCAUUUCCGUUUGCGCUCGUUCCCGUC-UCUACAGGC------------------- ((..(((((..((..((.....((((((((....)))).)))).............((((((........))))))))..)).))))).-...))....------------------- ( -23.60, z-score = 0.54, R) >droWil1.scaffold_181071 1054887 112 + 1211509 ----AAGUGAUGGGGCAAG-CUUU-UGUGCAACCGCGCAACAAUCAUUUUUGCACCGCAAAUUCAUUUCCGUUUGCGCUCGUUUUCACGAUGCAUUCAUUCACCGUCAACGAAUAAGU ----...((((((.(((((-(..(-(((((....))))))........(((((...))))).........))))))((((((....)))).)).........)))))).......... ( -29.90, z-score = -1.31, R) >consensus ____GUGUGU_GCGUCGAGUCCUU_UGUGCAACCGCACAACAAUCAUUUCGGCACCGCAAAUUCAUUUCCGUUUGCGCUCGUUUUUGCU_CACAUCAGC___________________ ......(((....((((((......(((((....)))))........))))))..(((((((........))))))).............)))......................... (-14.90 = -14.74 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:32 2011