| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,478,000 – 21,478,107 |
| Length | 107 |
| Max. P | 0.680294 |

| Location | 21,478,000 – 21,478,107 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 65.39 |
| Shannon entropy | 0.63265 |
| G+C content | 0.41107 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -12.53 |
| Energy contribution | -12.57 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
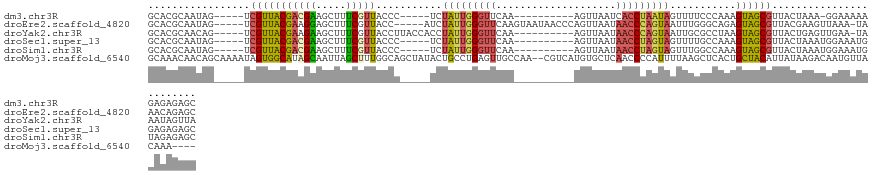
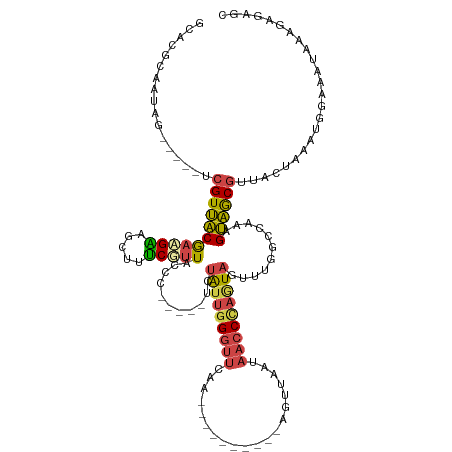
>dm3.chr3R 21478000 107 + 27905053 GCACGCAAUAG-----UCGUUACGACGAAGCUUUCGUUACCC-----UCUAUUGGGUUCAA----------AGUUAAUCACCUAAUAGUUUUCCCAAAGUAGCGUUACUAAA-GGAAAAAGAGAGAGC ..........(-----(((...))))...((((((.......-----.(((((((((....----------........)))))))))((((((...((((....))))...-))))))...)))))) ( -24.60, z-score = -1.59, R) >droEre2.scaffold_4820 3905901 117 - 10470090 GCACGCAAUAG-----UCGUUACGAAGGAGCUUUCGUUACC-----AUCUAUUGGGUUCAAGUAAUAACCCAGUUAAUAACCCAGUAAUUUGGGCAGAGUAGCGUUACGAAGUUAAA-UAAACAGAGC ....((.....-----((((.((((((....))))((((((-----((..((((((((........))))))))..))..(((((....)))))..).))))))).)))).(((...-..)))...)) ( -27.00, z-score = -1.64, R) >droYak2.chr3R 3836503 112 - 28832112 GCACGCAACAG-----UCGUUACGAAGAAGCUUUCGUUACCUUACCACCUAUUGGGUUCAA----------AGUUAAUAACCCAGUAAUUGCGCCUAAGUAGCGUUACUGAGUUGAA-UAAAUAGUUA ........(((-----(((((((..((..((....((......))....(((((((((...----------.......)))))))))...))..))..))))))..)))).......-.......... ( -21.80, z-score = -0.00, R) >droSec1.super_13 264862 108 + 2104621 GCACGCAAUAG-----UCGUUACGACGAAGCUUUCGUUACCC-----UCUAUUGGGUUCAA----------AGUUAAUAACCUAGUAGUUUUGCCAAAGUAGCGUUACUAAAUGGAAAUGGAGAGAGC ..........(-----(((...))))...(((((((..((((-----......)))).)..----------.........((...((((..(((.......)))..))))...)).......)))))) ( -24.60, z-score = -0.50, R) >droSim1.chr3R 21306353 108 + 27517382 GCACGCAAUAG-----UCGUUACGACGAAGCUUUCGUUACCC-----UCUAUUGGGUUCAA----------AGUUAAUAACCUAGUAGUUUGGCCAAAGUAGCGUUACUAAAUGGAAAUGUAGAGAGC ((((((....(-----(((...))))...(((((.((((...-----.((((((((((...----------.......))))))))))..)))).))))).))))..(((.((....)).)))...)) ( -26.00, z-score = -0.91, R) >droMoj3.scaffold_6540 8417251 122 - 34148556 GCAAACAACAGCAAAAUAGUGGCAUAGCAAUUAGCUUUGGCAGCUAUACUGCCUGAGUUGCCAA--CGUCAUGUGCUCAACCCCAUUUUAAGCUCACUGCUACAUUAUAAGACAAUGUUACAAA---- ((........))......(((((((((((....((.(((((((((..........)))))))))--.))....)))).............(((.....))).............)))))))...---- ( -27.90, z-score = -0.28, R) >consensus GCACGCAAUAG_____UCGUUACGAAGAAGCUUUCGUUACCC_____UCUAUUGGGUUCAA__________AGUUAAUAACCCAGUAGUUUGGCCAAAGUAGCGUUACUAAAUGGAAAUAAAGAGAGC .................(((((((((((.....)))))...........(((((((((....................)))))))))...........))))))........................ (-12.53 = -12.57 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:44 2011