
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,454,826 – 21,454,942 |
| Length | 116 |
| Max. P | 0.522415 |

| Location | 21,454,826 – 21,454,942 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 135 |
| Reading direction | reverse |
| Mean pairwise identity | 67.59 |
| Shannon entropy | 0.60922 |
| G+C content | 0.49389 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -14.48 |
| Energy contribution | -13.96 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
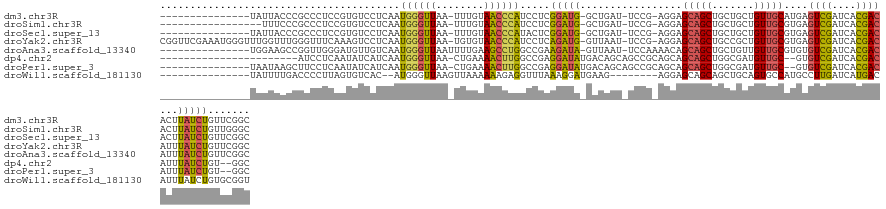
>dm3.chr3R 21454826 116 - 27905053 ---------------UAUUACCCGCCCUCCGUGUCCUCAAUGGGUUAA-UUUGUAACCCAUCCUCGGAUG-GCUGAU-UCCG-AGGAGCAGCUGCUGCUGUUGCAUGAGUCGAUCACGACACUUAUCUGUUCGGC ---------------........(((...(((((.....((((((((.-....))))))))(((((((..-......-))))-)))((((((....))))))))))).((((....))))............))) ( -40.60, z-score = -2.73, R) >droSim1.chr3R 21282675 114 - 27517382 -----------------UUUCCCGCCCUCCGUGUCCUCAAUGGGUUAA-UUUGUAACCCAUCCUCGGAUG-GCUGAU-UCCG-AGGAGCAGCUGCUGCUGUUGCGUGAGUCGAUCACGACACUUAUCUGUUGGGC -----------------......((((..((((((.....(((((((.-....)))))))((((((((..-......-))))-))))(((((.......)))))((((.....)))))))))......)..)))) ( -43.80, z-score = -3.18, R) >droSec1.super_13 241778 116 - 2104621 ---------------UAUUACCCGCCCUCCGUGUCCUCAAUGGGUUAA-UUUGUAACCCAUACUCGGAUG-GCUGAU-UCCG-AGGAGCAGCUGCUGCUGUUGCGUGAGUCGAUCACGACACUUAUCUGUUCGGC ---------------........(((.((((.((.....((((((((.-....)))))))))).)))).)-))....-.(((-((.((((((....)))).(((((((.....))))).)).....)).))))). ( -40.50, z-score = -2.56, R) >droYak2.chr3R 3812722 131 + 28832112 CGGUUCGAAAUGGGUUUGGUUUGGGUUUCAAAGUCCUCAAUGGGUUAA-UGUGUAACCCAUCCUCAGAUG-GUUAAU-UCCG-AGGAGCAGCUGCCGCUGUUGCGUGAGUCGAUCACGACAUUUAUCUGUUCGGC (((....(((((.((((((..((((((.((.(..((.....))..)..-..)).))))))..).)))))(-((..((-((..-.(.((((((....)))))).)..))))..)))....)))))..)))...... ( -36.90, z-score = -0.08, R) >droAna3.scaffold_13340 3711332 118 - 23697760 ---------------UGGAAGCCGGUUGGGAUGUUGUCAAUGGGUUAAUUUUGAAGCCUGGCCGAAGAUA-GUUAAU-UCCAAAACAGCAGCUGCUGUUGUUGCGUGUGUCGAUCACGACAUUUAUCUGUUCGGC ---------------.....((((((..((......((((..........))))..))..)))(((((((-(.....-.......(((((((....)))))))...((((((....))))))))))))..))))) ( -33.40, z-score = -0.54, R) >dp4.chr2 7139309 107 + 30794189 -----------------------AUCCUCAAUAUCAUCAAUGGGUUAA-CUGAAAACUUGGCCGAGGAUAUGACAGCAGCCGCAGCAGCAGCUGGCGAUGUUGC--GUGUCGAUCACGACAUUUAUCUGU--GGC -----------------------((((((.....((....))((((((-.(....).)))))))))))).........(((((((..(((((.......)))))--((((((....))))))....))))--))) ( -36.00, z-score = -1.75, R) >droPer1.super_3 945852 115 + 7375914 ---------------UAAUAAGCUUCCUCAAUAUCAUCAAUGGGUUAA-CUGAAAACUUGGCCGAGGAUAUGACAGCAGCCGCAGCAGCAGCUGGCGAUGUUGC--GUGUCGAUCACGACAUUUAUCUGU--GGC ---------------......((((((((.....((....))((((((-.(....).)))))))))))......))).(((((((..(((((.......)))))--((((((....))))))....))))--))) ( -36.70, z-score = -1.65, R) >droWil1.scaffold_181130 2065164 110 + 16660200 ---------------UAUUUUGACCCCUUAGUGUCAC--AUGGGUUAAGUUAAAAAAGAGGUUUAAAGGAUGAAG--------AGGAGCAGCAGCUGCAGUGCCAUGCCUUGAUCAUGACAUUUAUCUGUGCGGU ---------------...((((((((...........--..))))))))...........((.....(((((((.--------.(.(((....))).)..((.((((.......)))).)))))))))..))... ( -18.82, z-score = 2.43, R) >consensus _______________UAUUACCCGCCCUCAAUGUCCUCAAUGGGUUAA_UUUGAAACCCAGCCUAGGAUA_GAUAAU_UCCG_AGGAGCAGCUGCUGCUGUUGCGUGAGUCGAUCACGACAUUUAUCUGUUCGGC ........................................((((((........)))))).....(((((.................(((((.......)))))....((((....))))...)))))....... (-14.48 = -13.96 + -0.51)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:39 2011