| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,453,133 – 21,453,223 |
| Length | 90 |
| Max. P | 0.936991 |

| Location | 21,453,133 – 21,453,223 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.26 |
| Shannon entropy | 0.50744 |
| G+C content | 0.53092 |
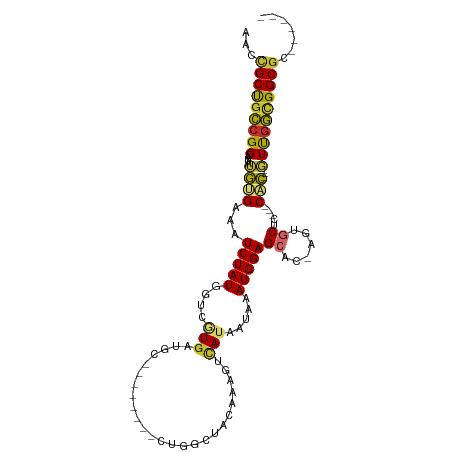
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -15.44 |
| Energy contribution | -15.23 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
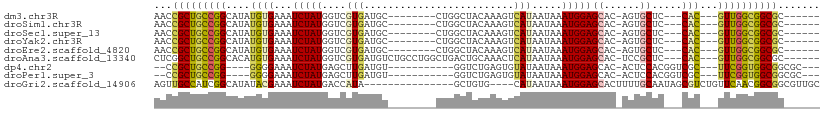
>dm3.chr3R 21453133 90 - 27905053 AACCGCUGCCGGCAUAUGUGAAAUCUAUGGUCGUGAUGC--------CUGGCUACAAAGUCAUAAUAAAUGGAGCAC-AGUGCUC---CAC---GUUGGCGGCGC------ ...((((((((((...(((((......((((((......--------.)))))).....))))).....((((((..-...))))---)).---)))))))))).------ ( -35.30, z-score = -2.81, R) >droSim1.chr3R 21281015 90 - 27517382 AACCGCUGCCGGCAUAUGUGAAAUCUAUGGUCGUGAUGC--------CUGGCUACAAAGUCAUAAUAAAUGGAGCAC-AGUGCUC---CAC---GUUGGCGGCGC------ ...((((((((((...(((((......((((((......--------.)))))).....))))).....((((((..-...))))---)).---)))))))))).------ ( -35.30, z-score = -2.81, R) >droSec1.super_13 240126 90 - 2104621 AACCGCUGCCGGCAUAUGUGAAAUCUAUGGUCGUGAUGC--------CUGGCUACAAAGUCAUAAUAAAUGGAGCAC-AGUGCUC---CAC---GUUGGCGGCGC------ ...((((((((((...(((((......((((((......--------.)))))).....))))).....((((((..-...))))---)).---)))))))))).------ ( -35.30, z-score = -2.81, R) >droYak2.chr3R 3810945 90 + 28832112 AACCGCUGCCGGCAUAUGUGAAAUCUAUGGUCGUGAUGC--------CUGGCUACAAAGUCAUAAUAAAUGGAGCAC-AGUGCUC---CAC---GUUGGCGGCGC------ ...((((((((((...(((((......((((((......--------.)))))).....))))).....((((((..-...))))---)).---)))))))))).------ ( -35.30, z-score = -2.81, R) >droEre2.scaffold_4820 3881257 90 + 10470090 AACCGCUGCCGGCAUAUGUGAAAUCUAUGGUCGUGAUGC--------CUGGCUACAAAGUCAUAAUAAAUGGAGCAC-AGUGCUC---CAC---GUUGGCGGCGC------ ...((((((((((...(((((......((((((......--------.)))))).....))))).....((((((..-...))))---)).---)))))))))).------ ( -35.30, z-score = -2.81, R) >droAna3.scaffold_13340 3709604 98 - 23697760 CUCGGCUGCCGGCACAUGUGAAAUCUAUGGUCGUGAUGUCUGCCUGGCUGACUGCAAACUCAUAAUAAAUGGAGCAC-UCCGCUC---CAC---GUUGGCGGCGC------ ....(((((((((...(((((.......(((((....(((.....))))))))......))))).....((((((..-...))))---)).---)))))))))..------ ( -33.82, z-score = -1.59, R) >dp4.chr2 7138085 87 + 30794189 --CCGCUGCCGG----GGGGAAAUCUAUGAGCUUGAUGU-----------GGUCUGAGUGUAUAAUAAAUGGAGCAC-ACUCCACGGUCGC---UUCGGUGGCGGCGC--- --((((..((((----((.((....((((.(((((((..-----------.))).)))).)))).....(((((...-.)))))...)).)---)))))..))))...--- ( -32.60, z-score = -1.71, R) >droPer1.super_3 944600 87 + 7375914 --CCGCUGCCGG----GGGGAAAUCUAUGAGCUUGAUGU-----------GGUCUGAGUGUAUAAUAAAUGGAGCAC-ACUCCACGGUCGC---UUCGGUGGCGGCGC--- --((((..((((----((.((....((((.(((((((..-----------.))).)))).)))).....(((((...-.)))))...)).)---)))))..))))...--- ( -32.60, z-score = -1.71, R) >droGri2.scaffold_14906 3002821 92 - 14172833 AGUUGCCAUCGGCAUAUACGAAAUCUAUGACCAUA---------------GCUGUG----CAUAAUAAAUGGAGCACUUUUGCAAUAGCGUCUGUUCAACGGCGGCGUUGC .((((((.(((.......)))......(((.((..---------------((((((----((..................))).)))))...)).)))..))))))..... ( -22.17, z-score = 0.56, R) >consensus AACCGCUGCCGGCAUAUGUGAAAUCUAUGGUCGUGAUGC________CUGGCUACAAAGUCAUAAUAAAUGGAGCAC_AGUGCUC___CAC___GUUGGCGGCGC______ ...(((((((((.....(((...(((((........................................)))))(((....))).....)))....)))))))))....... (-15.44 = -15.23 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:38 2011