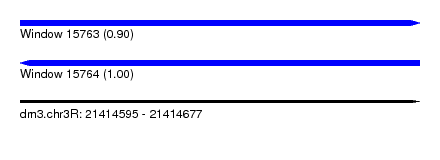
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,414,595 – 21,414,677 |
| Length | 82 |
| Max. P | 0.999610 |

| Location | 21,414,595 – 21,414,677 |
|---|---|
| Length | 82 |
| Sequences | 15 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 78.41 |
| Shannon entropy | 0.49487 |
| G+C content | 0.43603 |
| Mean single sequence MFE | -18.56 |
| Consensus MFE | -13.19 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
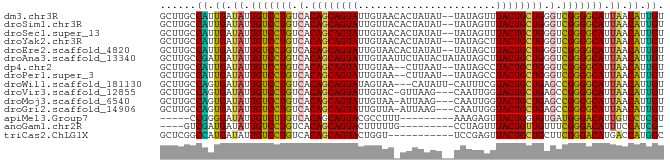
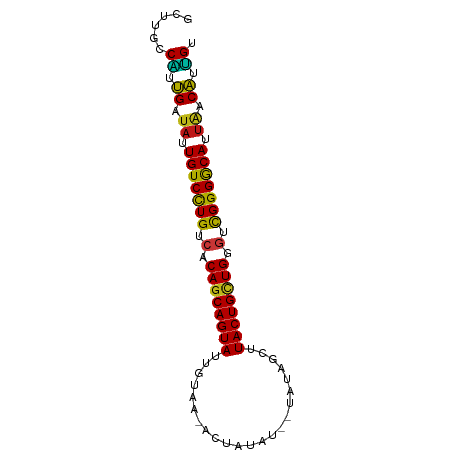
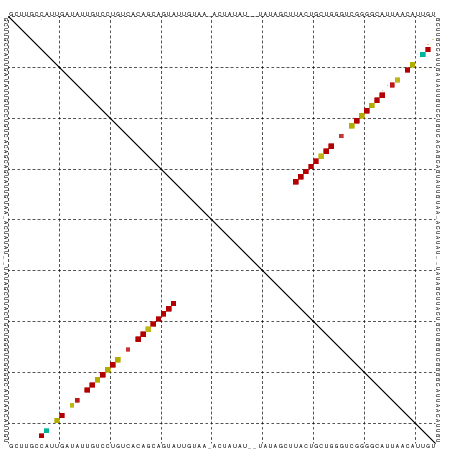
>dm3.chr3R 21414595 82 + 27905053 ACAAUGUUAAUGCCCCGACCCAGCAGUAAACUAUA--AUAUAGUGUUACAAUACUGCUGUGACAGGACAAUAUCAAUGGCAAGC ..........((((((...(((((((((.(((((.--..))))).......)))))))).)...))...........))))... ( -16.50, z-score = -0.62, R) >droSim1.chr3R 21243665 82 + 27517382 ACAAUGUUAAUGCCCCGACCCAGCAGUAAACUAUA--AUAUAGUGUAACAAUACUGCUGUGACAGGACAAUAUCAAUGGCAAGC ..........((((((...(((((((((.(((((.--..))))).......)))))))).)...))...........))))... ( -16.50, z-score = -0.75, R) >droSec1.super_13 202533 82 + 2104621 ACAAUGUUAAUGCCCCGACCCAGCAGUAAACUAUA--AUAUAGUGUUACAAUACUGCUGUGACAGGACAAUAUCAAUGGCAAGC ..........((((((...(((((((((.(((((.--..))))).......)))))))).)...))...........))))... ( -16.50, z-score = -0.62, R) >droYak2.chr3R 3767475 82 - 28832112 ACAAUGUUAAUGCCCCGACCCAGCAGUAAGCUAUA--AUAUAGUGUUACAAUACUGCUGUGACAGGACAAUAUCAAUGGCAAGC ..........((((((...(((((((((.(((((.--..))))).......)))))))).)...))...........))))... ( -16.80, z-score = -0.33, R) >droEre2.scaffold_4820 3842177 82 - 10470090 ACAAUGUUAAUGCCCCGACCCAGCAGUAAGCUAUA--AUAUAGUGUUACAAUACUGCUGUGACAGGACAAUAUCAAUGGCAAGC ..........((((((...(((((((((.(((((.--..))))).......)))))))).)...))...........))))... ( -16.80, z-score = -0.33, R) >droAna3.scaffold_13340 3670438 84 + 23697760 ACAAUGUUAAUGCCCCGACCCAGCAGUAAGCUAUAUAGUAUAGAAUUACAAUACUGCUGUGACAGGACAAUAUCAUCGGCAAGC ..........((((((...(((((((((..(((((...)))))........)))))))).)...))...........))))... ( -16.10, z-score = -0.51, R) >dp4.chr2 21158179 80 + 30794189 ACAAUGUUAAUGCCCCGACCCAGCAGUAGGCUAUA--AUUAAG--UUACAAUACUGCUGUGACAGGACAAUAUCAAUGGCAAGC ..........((((((...(((((((((((((...--....))--))....)))))))).)...))...........))))... ( -16.70, z-score = -0.34, R) >droPer1.super_3 3932668 80 + 7375914 ACAAUGUUAAUGCCCCGACCCAGCAGUAGGCUAUA--AUUAAG--UUACAAUACUGCUGUGACAGGACAAUAUCAAUGGCAAGC ..........((((((...(((((((((((((...--....))--))....)))))))).)...))...........))))... ( -16.70, z-score = -0.34, R) >droWil1.scaffold_181130 2021406 80 - 16660200 ACAAUGUUAAUGCCCCGGCUCAGCAGUACGAAAUG-AAUAUG---UUACUAUACUGCUGUGACAGGACAAUAUCACUGGCAAGC ..........((((((..(.((((((((...(((.-.....)---))....)))))))).)...))...........))))... ( -16.70, z-score = -0.01, R) >droVir3.scaffold_12855 6810278 80 + 10161210 ACAAUGUUAAUGCCCCGGCUCAGCAGUACCAAUUG---CUUAAC-GUACAAUACUGCUGUGACAGGACAAUAUCACUGGCAAGC ..........((((((..(.((((((((.....((---(.....-)))...)))))))).)...))...........))))... ( -17.80, z-score = -0.06, R) >droMoj3.scaffold_6540 8356923 80 - 34148556 ACAAUGUUAAUGCCCCGGCUCAGCAGUACCAAUUG---CUUAAU-UUACAAUACUGCUGUGACAGGACAAUAUCACUGGCAAGC ..........((((((..(.((((((((....(((---......-...))))))))))).)...))...........))))... ( -16.30, z-score = 0.10, R) >droGri2.scaffold_14906 2964728 80 + 14172833 ACAAUGUUAAUGCCCCGGCUCAGCAGUACCAAUUG---CUUAAU-UAACAAUACUGCUGUGACAGGACAAUAUCACUGGCAAGC ....(((((((((....))..((((((....))))---))..))-)))))....((((((((.(......).)))).))))... ( -17.60, z-score = -0.39, R) >apiMel3.Group7 2292704 70 - 9974240 ACGAGGACAAUGUCCCAUCACACCAGUAACUCUUU---------AAAGGCGUACUGCUGUGACAAGACAAUAUCCCCGG----- .((.(((.(.((((...(((((.(((((..(((..---------..)))..))))).)))))...)))).).))).)).----- ( -21.70, z-score = -3.59, R) >anoGam1.chr2R 49484070 70 - 62725911 -CGAUGGAAAUGUCCCGAAACAACAGUAAACUAGG---------CAAAAAGUACUGCUGUGACAGGACAAUAUCAUCGAC---- -((((((.(.(((((.(..(((.((((..(((...---------.....))))))).)))..).))))).).))))))..---- ( -20.70, z-score = -4.11, R) >triCas2.ChLG1X 2776360 73 - 8109244 GCCAUGGUCAUGUCCCGAAGCAGCAGUAACUCGGA-----------ACCAGUACUGCUGUGACAGGACAAUAUCAUGGCCGAGC ((((((((..(((((.(..(((((((((.((.(..-----------..))))))))))))..).)))))..))))))))..... ( -35.00, z-score = -5.41, R) >consensus ACAAUGUUAAUGCCCCGACCCAGCAGUAAGCUAUA__AUAUAGU_UUACAAUACUGCUGUGACAGGACAAUAUCAAUGGCAAGC .((.((.((.((.((.(.(.((((((((.......................)))))))).).).)).)).)).)).))...... (-13.19 = -13.43 + 0.23)

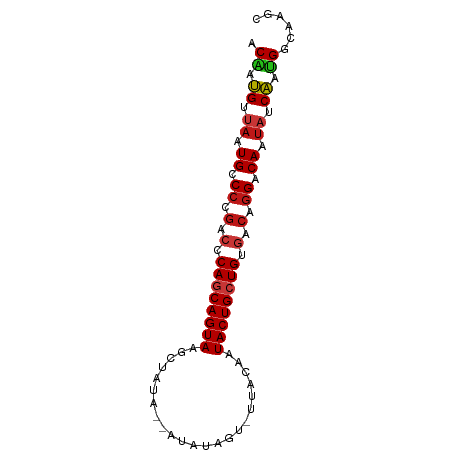
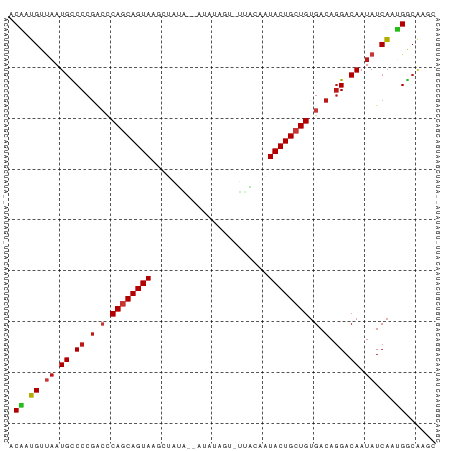
| Location | 21,414,595 – 21,414,677 |
|---|---|
| Length | 82 |
| Sequences | 15 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 78.41 |
| Shannon entropy | 0.49487 |
| G+C content | 0.43603 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -23.55 |
| Energy contribution | -22.98 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.41 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21414595 82 - 27905053 GCUUGCCAUUGAUAUUGUCCUGUCACAGCAGUAUUGUAACACUAUAU--UAUAGUUUACUGCUGGGUCGGGGCAUUAACAUUGU ......((.((.((.(((((((.(.((((((((((((((.......)--))))))..))))))).).))))))).)).)).)). ( -27.10, z-score = -3.01, R) >droSim1.chr3R 21243665 82 - 27517382 GCUUGCCAUUGAUAUUGUCCUGUCACAGCAGUAUUGUUACACUAUAU--UAUAGUUUACUGCUGGGUCGGGGCAUUAACAUUGU ......((.((.((.(((((((.(.((((((((.......(((((..--.))))).)))))))).).))))))).)).)).)). ( -25.90, z-score = -2.65, R) >droSec1.super_13 202533 82 - 2104621 GCUUGCCAUUGAUAUUGUCCUGUCACAGCAGUAUUGUAACACUAUAU--UAUAGUUUACUGCUGGGUCGGGGCAUUAACAUUGU ......((.((.((.(((((((.(.((((((((((((((.......)--))))))..))))))).).))))))).)).)).)). ( -27.10, z-score = -3.01, R) >droYak2.chr3R 3767475 82 + 28832112 GCUUGCCAUUGAUAUUGUCCUGUCACAGCAGUAUUGUAACACUAUAU--UAUAGCUUACUGCUGGGUCGGGGCAUUAACAUUGU ......((.((.((.(((((((.(.((((((((((((((.......)--)))))..)))))))).).))))))).)).)).)). ( -27.00, z-score = -2.75, R) >droEre2.scaffold_4820 3842177 82 + 10470090 GCUUGCCAUUGAUAUUGUCCUGUCACAGCAGUAUUGUAACACUAUAU--UAUAGCUUACUGCUGGGUCGGGGCAUUAACAUUGU ......((.((.((.(((((((.(.((((((((((((((.......)--)))))..)))))))).).))))))).)).)).)). ( -27.00, z-score = -2.75, R) >droAna3.scaffold_13340 3670438 84 - 23697760 GCUUGCCGAUGAUAUUGUCCUGUCACAGCAGUAUUGUAAUUCUAUACUAUAUAGCUUACUGCUGGGUCGGGGCAUUAACAUUGU ......(((((.((.(((((((.(.(((((((((((((...........)))))..)))))))).).))))))).)).))))). ( -28.50, z-score = -3.15, R) >dp4.chr2 21158179 80 - 30794189 GCUUGCCAUUGAUAUUGUCCUGUCACAGCAGUAUUGUAA--CUUAAU--UAUAGCCUACUGCUGGGUCGGGGCAUUAACAUUGU ......((.((.((.(((((((.(.((((((((((((((--.....)--)))))..)))))))).).))))))).)).)).)). ( -27.30, z-score = -2.91, R) >droPer1.super_3 3932668 80 - 7375914 GCUUGCCAUUGAUAUUGUCCUGUCACAGCAGUAUUGUAA--CUUAAU--UAUAGCCUACUGCUGGGUCGGGGCAUUAACAUUGU ......((.((.((.(((((((.(.((((((((((((((--.....)--)))))..)))))))).).))))))).)).)).)). ( -27.30, z-score = -2.91, R) >droWil1.scaffold_181130 2021406 80 + 16660200 GCUUGCCAGUGAUAUUGUCCUGUCACAGCAGUAUAGUAA---CAUAUU-CAUUUCGUACUGCUGAGCCGGGGCAUUAACAUUGU ......(((((.((.(((((((.(.(((((((((.....---......-......))))))))).).))))))).)).))))). ( -28.13, z-score = -3.38, R) >droVir3.scaffold_12855 6810278 80 - 10161210 GCUUGCCAGUGAUAUUGUCCUGUCACAGCAGUAUUGUAC-GUUAAG---CAAUUGGUACUGCUGAGCCGGGGCAUUAACAUUGU ......(((((.((.(((((((.(.(((((((((((...-(.....---)...))))))))))).).))))))).)).))))). ( -29.70, z-score = -2.70, R) >droMoj3.scaffold_6540 8356923 80 + 34148556 GCUUGCCAGUGAUAUUGUCCUGUCACAGCAGUAUUGUAA-AUUAAG---CAAUUGGUACUGCUGAGCCGGGGCAUUAACAUUGU ......(((((.((.(((((((.(.((((((((((((..-.....)---))....))))))))).).))))))).)).))))). ( -29.50, z-score = -3.03, R) >droGri2.scaffold_14906 2964728 80 - 14172833 GCUUGCCAGUGAUAUUGUCCUGUCACAGCAGUAUUGUUA-AUUAAG---CAAUUGGUACUGCUGAGCCGGGGCAUUAACAUUGU ......(((((.((.(((((((.(.(((((((((((((.-....))---))....))))))))).).))))))).)).))))). ( -30.40, z-score = -3.33, R) >apiMel3.Group7 2292704 70 + 9974240 -----CCGGGGAUAUUGUCUUGUCACAGCAGUACGCCUUU---------AAAGAGUUACUGGUGUGAUGGGACAUUGUCCUCGU -----.((((((((.(((((..(((((.(((((.((((..---------..)).))))))).)))))..))))).)))))))). ( -38.20, z-score = -7.28, R) >anoGam1.chr2R 49484070 70 + 62725911 ----GUCGAUGAUAUUGUCCUGUCACAGCAGUACUUUUUG---------CCUAGUUUACUGUUGUUUCGGGACAUUUCCAUCG- ----..(((((..(.(((((((..(((((((((..((...---------...))..)))))))))..))))))).)..)))))- ( -28.00, z-score = -6.61, R) >triCas2.ChLG1X 2776360 73 + 8109244 GCUCGGCCAUGAUAUUGUCCUGUCACAGCAGUACUGGU-----------UCCGAGUUACUGCUGCUUCGGGACAUGACCAUGGC .....((((((....(((((((...(((((((((((..-----------..).)).))))))))...)))))))....)))))) ( -33.30, z-score = -4.19, R) >consensus GCUUGCCAUUGAUAUUGUCCUGUCACAGCAGUAUUGUAA_ACUAUAU__UAUAGCUUACUGCUGGGUCGGGGCAUUAACAUUGU ......((.((.((.(((((((.(.((((((((.......................)))))))).).))))))).)).)).)). (-23.55 = -22.98 + -0.57)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:33 2011