| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,396,986 – 21,397,082 |
| Length | 96 |
| Max. P | 0.821733 |

| Location | 21,396,986 – 21,397,082 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.94 |
| Shannon entropy | 0.53262 |
| G+C content | 0.47435 |
| Mean single sequence MFE | -30.21 |
| Consensus MFE | -11.49 |
| Energy contribution | -10.97 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
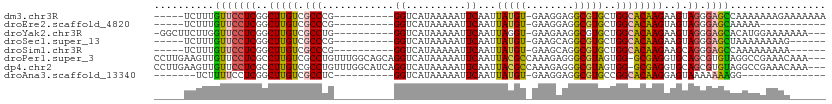
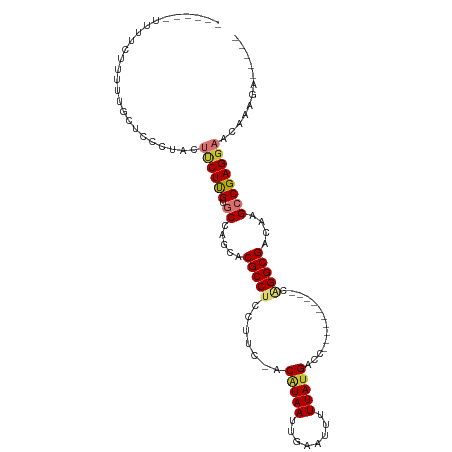
>dm3.chr3R 21396986 96 + 27905053 -----UCUUUGUUCCUCGGCUUGUCGCCCG----------GGUCAUAAAAAUUCAAUUAUGU-GAAGGAGGCGUGCUGGCACAAGAAGUAGGGAGCCAAAAAAAGAAAAAAA -----(((((((((((..(((((((..((.----------..(((((.((......)).)))-)).)).)))(((....)))...)))).)))))).....)))))...... ( -22.70, z-score = -0.30, R) >droEre2.scaffold_4820 3824543 85 - 10470090 -----UCUUUGUUCCUCGGCUUGUCGCCCG----------GGUCAUAAAAAUUCAAUUAUGU-GAAGGAGGCGUGCUGGCACAAGUAGUAGGGAGCAAAAA----------- -----..(((((((((..((((((.(((((----------.(((.......((((......)-)))...))).))..)))))))))....)))))))))..----------- ( -26.50, z-score = -1.94, R) >droYak2.chr3R 3749190 97 - 28832112 -GGCUUCUUGGUUCCUCGGCUUGUCGCCUG----------GGUCAUAAAAAUUCAAUUAGGU-GAAGAAGGCGUGCUGGCACAAGAAGUAGGGAGCACAUGGAAAAAAA--- -.((((((((.(((....(((.(((((((.----------..((((..((......))..))-))...))))).)).)))....))).)))))))).............--- ( -30.80, z-score = -1.98, R) >droSec1.super_13 184977 90 + 2104621 -----UCUUUGUUCCUCGGCUUGUCGCCCG----------GGUCAUAAAAAUUCAAUUAUGU-GAAGCAGGCGUGCUGGCACAAGAAGUAGGGAGCUAAAAAAAAG------ -----.....((((((..((((...(((.(----------..(((((.((......)).)))-))..).)))(((....)))...)))).))))))..........------ ( -22.40, z-score = -0.51, R) >droSim1.chr3R 21225634 90 + 27517382 -----UCUUUGUUCCUCGGCUUGUCGCCCG----------GGUCAUAAAAAUUCAAUUAUGU-GAAGCAGGCGUGCUGGCACAAGAAGCAGGGAGCCAAAAAAAAA------ -----((((((((.((.((((.((((((.(----------..(((((.((......)).)))-))..).)))).)).))).).)).))))))))............------ ( -24.30, z-score = -0.54, R) >droPer1.super_3 3910207 108 + 7375914 CCUUGAAGUUGUUCCUCGCCUUGUCGCCUGUUUGGCAGCAGGUCAUAAAAAUUCAAUUACGCCAAAGAGGGCGUAGUGG-GCGAGGUGCAGCGUGUAGGCCGAAACAAA--- (((....(((((.(((((((((((.((((((......)))))).)))).......((((((((......)))))))).)-)))))).)))))....)))..........--- ( -45.50, z-score = -4.09, R) >dp4.chr2 21135675 108 + 30794189 CCUUGAAGUUGUUCCUCGCCUUGUCGCCUGUUUGGCAUCAGGUCAUAAAAAUUCAAUUACGCCAAAGAGGGCGUAGUGG-GCGAGGUGCAGCGUGUAGGCCGAAACAAA--- (((....(((((.(((((((((((.(((((........))))).)))).......((((((((......)))))))).)-)))))).)))))....)))..........--- ( -43.70, z-score = -3.94, R) >droAna3.scaffold_13340 3652310 80 + 23697760 -------UCUUUUCCUCGGCUUGUCGCCUC----------GGUCAUAAAAAUUCAAUUAUGU-GAAGGAGGCGUGCCGGCACAAGGAGUAAAAAAAGG-------------- -------....(((((.((((.((((((((----------..(((((.((......)).)))-))..)))))).)).))).).)))))..........-------------- ( -25.80, z-score = -2.98, R) >consensus _____UCUUUGUUCCUCGGCUUGUCGCCCG__________GGUCAUAAAAAUUCAAUUAUGU_GAAGAAGGCGUGCUGGCACAAGAAGUAGGGAGCAAAAAAAAAA______ ..........(((((((..(((((.(((............((..........))...(((((........)))))..))))))))..).)).))))................ (-11.49 = -10.97 + -0.51)
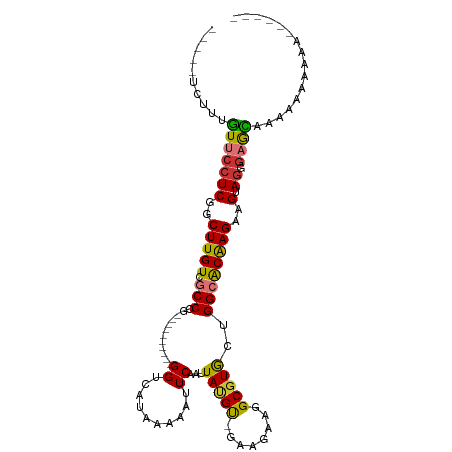
| Location | 21,396,986 – 21,397,082 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.94 |
| Shannon entropy | 0.53262 |
| G+C content | 0.47435 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -10.72 |
| Energy contribution | -11.06 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21396986 96 - 27905053 UUUUUUUCUUUUUUUGGCUCCCUACUUCUUGUGCCAGCACGCCUCCUUC-ACAUAAUUGAAUUUUUAUGACC----------CGGGCGACAAGCCGAGGAACAAAGA----- ...((((..((((((((((...........(((....)))((((.....-.(((((........)))))...----------.))))....))))))))))..))))----- ( -20.90, z-score = -1.23, R) >droEre2.scaffold_4820 3824543 85 + 10470090 -----------UUUUUGCUCCCUACUACUUGUGCCAGCACGCCUCCUUC-ACAUAAUUGAAUUUUUAUGACC----------CGGGCGACAAGCCGAGGAACAAAGA----- -----------((((((.(((......((((((....).(((((.....-.(((((........)))))...----------.))))))))))....))).))))))----- ( -17.70, z-score = -1.16, R) >droYak2.chr3R 3749190 97 + 28832112 ---UUUUUUUCCAUGUGCUCCCUACUUCUUGUGCCAGCACGCCUUCUUC-ACCUAAUUGAAUUUUUAUGACC----------CAGGCGACAAGCCGAGGAACCAAGAAGCC- ---...........(((((...(((.....)))..)))))..(((((((-(......)))..........((----------..(((.....)))..)).....)))))..- ( -18.70, z-score = -0.78, R) >droSec1.super_13 184977 90 - 2104621 ------CUUUUUUUUAGCUCCCUACUUCUUGUGCCAGCACGCCUGCUUC-ACAUAAUUGAAUUUUUAUGACC----------CGGGCGACAAGCCGAGGAACAAAGA----- ------..............(((....((((((((((((....))))..-.(((((........)))))...----------..))).)))))...)))........----- ( -18.50, z-score = -0.63, R) >droSim1.chr3R 21225634 90 - 27517382 ------UUUUUUUUUGGCUCCCUGCUUCUUGUGCCAGCACGCCUGCUUC-ACAUAAUUGAAUUUUUAUGACC----------CGGGCGACAAGCCGAGGAACAAAGA----- ------...((((((((((...((((.........))))((((((....-.(((((........)))))...----------))))))...))))))))))......----- ( -23.50, z-score = -1.35, R) >droPer1.super_3 3910207 108 - 7375914 ---UUUGUUUCGGCCUACACGCUGCACCUCGC-CCACUACGCCCUCUUUGGCGUAAUUGAAUUUUUAUGACCUGCUGCCAAACAGGCGACAAGGCGAGGAACAACUUCAAGG ---.(((((.((((......))))..((((((-(...((((((......)))))).................((.((((.....)))).)).))))))))))))........ ( -33.30, z-score = -2.48, R) >dp4.chr2 21135675 108 - 30794189 ---UUUGUUUCGGCCUACACGCUGCACCUCGC-CCACUACGCCCUCUUUGGCGUAAUUGAAUUUUUAUGACCUGAUGCCAAACAGGCGACAAGGCGAGGAACAACUUCAAGG ---.(((((.((((......))))..((((((-(.....((((...((((((((......((....))......))))))))..))))....))))))))))))........ ( -34.10, z-score = -3.02, R) >droAna3.scaffold_13340 3652310 80 - 23697760 --------------CCUUUUUUUACUCCUUGUGCCGGCACGCCUCCUUC-ACAUAAUUGAAUUUUUAUGACC----------GAGGCGACAAGCCGAGGAAAAGA------- --------------...........((((((.((..(..((((((....-.(((((........)))))...----------)))))).)..)))))))).....------- ( -22.70, z-score = -3.06, R) >consensus ______UUUUCUUUUUGCUCCCUACUUCUUGUGCCAGCACGCCUCCUUC_ACAUAAUUGAAUUUUUAUGACC__________CAGGCGACAAGCCGAGGAACAAAGA_____ .........................((((((.((.....(((((.......(((((........)))))..............)))))....))))))))............ (-10.72 = -11.06 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:30 2011