
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,386,290 – 21,386,385 |
| Length | 95 |
| Max. P | 0.757895 |

| Location | 21,386,290 – 21,386,385 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 66.49 |
| Shannon entropy | 0.63666 |
| G+C content | 0.41411 |
| Mean single sequence MFE | -20.01 |
| Consensus MFE | -8.99 |
| Energy contribution | -10.61 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21386290 95 + 27905053 -CUCCAGUGGAGCUGGGCCUCGAAUUUUAACCCCAAGGUGACCGAGAGGUGUAGUUUAACGAAGAAUAAACAUAAAAAAAAA-------AUACCAUUAAUAUG -.....((.((((((.(((((........(((....)))......))))).)))))).))......................-------.............. ( -19.14, z-score = -1.30, R) >droAna3.scaffold_13340 3642053 91 + 23697760 CGUAAGC-CAAGCCAAACCGCGAGUUA-ACCCUCAAGGUGAACCCAGG---UGAGUGGGGGAAAUGAAGAAUUAAAUAAUAAU-------UGGAAAUAAAAUG ......(-(((((......))......-.((((((......((....)---)...)))))).....................)-------))).......... ( -14.70, z-score = 0.44, R) >droEre2.scaffold_4820 3813634 91 - 10470090 -CUCCAGUGGAGCUGGGCUUCGAGUUUUAACCCCAAGGUGACCGUGAGGUGUAGCUUAACGAGGAAUAAACAUAAAAACA-----------ACAAUUAAAACG -.(((.((.((((((.((((((.......(((....))).....)))))).)))))).))..)))...............-----------............ ( -22.90, z-score = -1.52, R) >droYak2.chr3R 3738133 102 - 28832112 -CUCCAGUGGAGCUGGGCCUCGAAUUUUAACCCCAAGGUGACCGUGAGGUGUAGUUUAACGAAGAGUGAACAUAAAAAAACAUAAAAAAAUACAAUUAAAAUG -(((..((.((((((.((((((.......(((....))).....)))))).)))))).))...)))..................................... ( -22.00, z-score = -1.79, R) >droSec1.super_13 174233 88 + 2104621 -CUCCAGUGGAGCUGGGCCUCGAAUUUUAACCCCAAAGUGA---GGAGGUGUAGUUCAACGAAGAAUAAACAUAAAAAA-----------UGUCAUUAAAAUG -....((((((((((.(((((..(((((......)))))..---.))))).))))))............((((.....)-----------)))))))...... ( -19.40, z-score = -1.48, R) >droSim1.chr3R 21214905 92 + 27517382 -CUCCAGUGGAGCUGGGCCUCGAAUUUUAACCCCAAGGUGACCGAGAGGUGUAGUUUAACGAAGAAUAAACAUAAAAAAA----------UGUCAUUAAAAUG -.....((.((((((.(((((........(((....)))......))))).)))))).)).........((((......)----------))).......... ( -19.54, z-score = -1.18, R) >droWil1.scaffold_181130 1990277 83 - 16660200 -GUCAGUUCGUGUGGGGAGGCGAGUU---CUCUCAAGGUGAACCACAC---CA--UGGGCAAAAGCCAG---UAAA-AAUGCU-------UGCAAUUAAAAUG -....(((((((((((((((.....)---)))))..((....)).)).---))--)))))....((.((---((..-..))))-------.)).......... ( -22.40, z-score = -1.14, R) >consensus _CUCCAGUGGAGCUGGGCCUCGAAUUUUAACCCCAAGGUGACCGAGAGGUGUAGUUUAACGAAGAAUAAACAUAAAAAAA__________UGCAAUUAAAAUG ......((.((((((.(((((........(((....)))......))))).)))))).))........................................... ( -8.99 = -10.61 + 1.62)

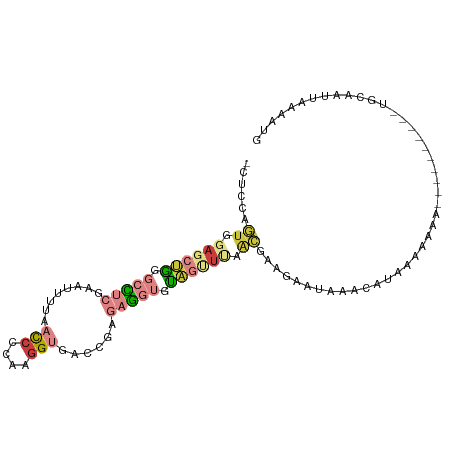
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:28 2011