
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,381,576 – 21,381,707 |
| Length | 131 |
| Max. P | 0.895469 |

| Location | 21,381,576 – 21,381,707 |
|---|---|
| Length | 131 |
| Sequences | 7 |
| Columns | 145 |
| Reading direction | reverse |
| Mean pairwise identity | 59.88 |
| Shannon entropy | 0.75050 |
| G+C content | 0.59609 |
| Mean single sequence MFE | -40.27 |
| Consensus MFE | -26.39 |
| Energy contribution | -24.87 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.52 |
| Mean z-score | -0.15 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21381576 131 - 27905053 ---CUCCCCCCCCCCCCAAAAAAAGGCCCCAACUCCAACUCC-------UUUCCCAU--UUUUUUCUCAGUUCCGCC--GUCUUACUCGCAGAGACUCCACUCGUUGGGGCGUUGGAGCGUUGUAGCCGGGUGAGUGGAACGAGA ---..................(((((..............))-------))).....--.....((((.(((((((.--(((((.......)))))..(((((((((.(((((....))))).))).)))))).))))))))))) ( -34.44, z-score = 0.55, R) >droSim1.chr3R 21210288 125 - 27517382 ----------CUCCCCCCAAAAAGG-CCCCAACUCCAACUCC-------UUUCCCAUUCUUUUUUUUCAGUUCCGCC--GUCUUACUCGCAGAGACUCCACUCGUUGGGGCGCUGGAGCGUUUUAGCCGAGCGAGCGGAACAGGG ----------.....(((..(((((-..............))-------))).................(((((((.--(((((.......)))))..(.(((((((((((((....))))))))).)))).).))))))).))) ( -36.84, z-score = -0.72, R) >droSec1.super_13 169634 136 - 2104621 CUACCCCCCCCCCCCCCCAAAAAGGCCCCCAACUCCAACUCC-------UUUCCCAUUCUUUUUUUUCAGUUCCGCC--GUCUUACUCGCAGAGACUCCACUCGUUGGGGCGUUGGAGCGUUAUAGCCGGGCGAGCGGAACGGGG ..............((((..(((((...............))-------))).................((((((((--((((..((......(.(((((..(((....))).)))))).....))..))))).))))))))))) ( -37.56, z-score = 0.39, R) >droEre2.scaffold_4820 3808943 135 + 10470090 --------CUCCCCCCUCGAAAAAGACCCCAACUCCUACUUCAACUCCAAUUCCUUUCCCAUUUUUUCAGUUCCGCC--GUCUUGCUCGCAGAGUCUCCACUCGUUGGGGCGUUGGAGCGUUAUAGCCGGGUGAGCGGAACGGAG --------((((......(((((((....................................))))))).((((((((--..((.(((......(.(((((..(((....))).)))))).....))).))..).))))))))))) ( -34.32, z-score = 0.59, R) >droAna3.scaffold_13340 3637315 142 - 23697760 CUCCUUCUCCGUUUCUUCAUUCCAGUUCCGCCGUCUUGCCUU---CUCAGUCACUUUCGGCUCAGCUCGGCCUAGCCUGGCCUGGCUCCACGCCACUCCACUCGUUGGGGCGUUGGAGCGUUAUAACUGGGUGAGCAUGGCAGGG ..(((...((((...(((((.((((((..((((...((.((.---...)).))....))))..(((..((((......))))..((((((((((...(((.....))))))).)))))))))..))))))))))).)))).))). ( -49.50, z-score = -1.23, R) >dp4.chr2 21115149 120 - 30794189 ----------------------CUGGCUCUCCCGCUCACCGGAUGGUUCUUUCCCUGCCUCCCCCCUCCACUCCGU---GAGCGACGACGACGUUUCCCACUCGUUGGGGCGUUCGAGCGUUAUAACUGGGUGAGCGGGGCAUGG ----------------------((.((((.(((((((((.((((((.....................))).)))))---))))((((.((((((..((((.....))))))).)))..))))......))).)))).))...... ( -44.60, z-score = -0.25, R) >droPer1.super_3 3889680 120 - 7375914 ----------------------CUGGCUCUCCCGCUCACCGGAUGGUUCUUUCCCUGCCUUCCCCCUCCACUCCGU---GAGCGACGACGACGUUUCCCACUCGUUGGGGCGUUCGAGCGUUAUAACUGGGUGAGCGGGGCAUGG ----------------------((.((((.(((((((((.((((((.....................))).)))))---))))((((.((((((..((((.....))))))).)))..))))......))).)))).))...... ( -44.60, z-score = -0.41, R) >consensus ________C_CCCCCCCCAAAAAAGGCCCCAACUCCAACUCC_______UUUCCCUUCCUUUUUUCUCAGUUCCGCC__GUCUUACUCGCAGAGACUCCACUCGUUGGGGCGUUGGAGCGUUAUAGCCGGGUGAGCGGAACAGGG .....................................................................(((((((...(((((.......)))))..(((((((((..((((....))))..)))).))))).))))))).... (-26.39 = -24.87 + -1.52)
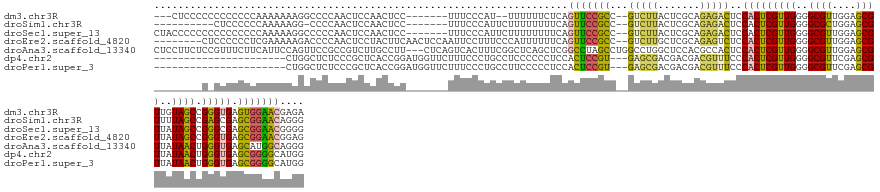

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:27 2011