
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,362,556 – 21,362,693 |
| Length | 137 |
| Max. P | 0.749004 |

| Location | 21,362,556 – 21,362,653 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.51 |
| Shannon entropy | 0.49899 |
| G+C content | 0.48797 |
| Mean single sequence MFE | -33.22 |
| Consensus MFE | -12.75 |
| Energy contribution | -12.46 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
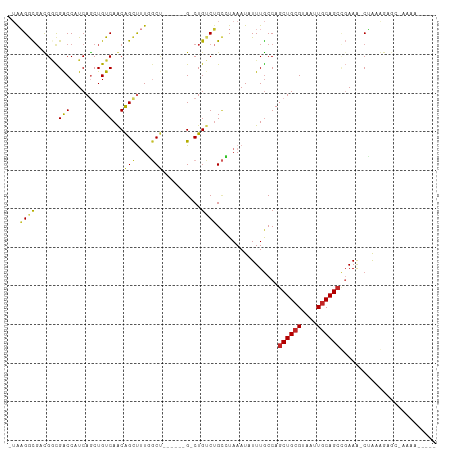
>dm3.chr3R 21362556 97 + 27905053 -UAAGGCGUCGGCGACCAUCAGCUGUCAGCAGCUUUGGCU------G-CUGUCUGCCUAAAUAUUUGCCAGCUGCGUAAUUGCAGCCGAAA-CUACGGAGCAAAAA----- -..(((((.(((((.(((..(((((....))))).))).)------)-))).).)))).....(((((..((((((....)))))).....-(....).)))))..----- ( -37.80, z-score = -2.40, R) >droSim1.chr3R 21190248 97 + 27517382 -UAAGGCGUCGGCGACCAUCAGCUGUCAGCAGCUUUGGCU------G-CUGUCUGCCUAAAUAUUUGCCAGCUGCGUAAUUGCAGCCGAAA-CUGCGGAGCAAAAA----- -..(((((.(((((.(((..(((((....))))).))).)------)-))).).)))).....((((((.((((((....))))))((...-...))).)))))..----- ( -37.60, z-score = -1.80, R) >droSec1.super_13 149050 97 + 2104621 -UAAGGCGUCGGCGACCAUCAGCUGUCAGCAGCUUUGGCU------G-CUGUCUGCCUAAAUAUUUGCCAGCUGCGUAAUUGCAGCCGAAA-CUACGGAGCAAAAA----- -..(((((.(((((.(((..(((((....))))).))).)------)-))).).)))).....(((((..((((((....)))))).....-(....).)))))..----- ( -37.80, z-score = -2.40, R) >droYak2.chr3R 3713107 98 - 28832112 -UAAGGCGUCGGCGACCAUCAGCUGUCAGCAGCUUUGGCU------G-CUGUCUGCCUAAAUAUUUGCCAGCUGCGUAAUUGCAGCCGAAA-CUGCGGUGGAAAAAA---- -..(((((.(((((.(((..(((((....))))).))).)------)-))).).)))).....((..((.((((((....)))))).(...-...)))..)).....---- ( -37.60, z-score = -1.88, R) >droEre2.scaffold_4820 3788108 102 - 10470090 -UAAGGCGUCGGCGACCAUCAGCUGUCAGCAGCUUUGGCU------G-CUGUCUGCCUAAAUAUUUGCCAGCUGCGUAAUUGCAGGCGAAA-CUAGGGAGGUAAAAAUAAA -..(((((.(((((.(((..(((((....))))).))).)------)-))).).)))).....((((((..(((((....))))).(....-...)...))))))...... ( -34.60, z-score = -1.08, R) >droAna3.scaffold_13340 3617790 101 + 23697760 -UGAGACUACGUCGGCCAUCAGCUGUCAACAGCUUUUGCC------G-CUGUCUGCGUAAAUAUUUGCCAGCUGCGUAAUUGCAGCCGAAAACUAGAGAGGCAGUAAAA-- -...(((...)))(((.(..(((((....)))))..))))------(-(((((((((........)))..((((((....))))))............)))))))....-- ( -31.40, z-score = -1.13, R) >dp4.chr2 21089582 97 + 30794189 -UGAGGCGAUGACGACCAUCAACUGUCAACAGCCUUUGCUUCUGCUG-CUGUCUGCGUAAAUAUUUGCCAGCUGCGUAAUUGCAGCUGAAACCGAAAAA------------ -...(((((((.....))))....((..(((((....((....)).)-))))..))..........)))(((((((....)))))))............------------ ( -28.90, z-score = -2.13, R) >droPer1.super_3 3863024 97 + 7375914 -UGAGGCGAUGACGACCAUCAACUGUCAACAGCCUUUGCUUCUGCUG-CUGUCUGCGUAAAUAUUUGCCAGCUGCGUAAUUGCAGCUGAAACCGAAAAA------------ -...(((((((.....))))....((..(((((....((....)).)-))))..))..........)))(((((((....)))))))............------------ ( -28.90, z-score = -2.13, R) >droVir3.scaffold_12855 3430426 99 - 10161210 UGAUGAUGAUGAUGACCAUCAGCUGUCAACGGUUCUGUCCCGAACUGUCUGACAGUGUAAAUAUUUGCCAGCUGCGCAAUUGCAGC--AAAACAAAAAAAA---------- ......(((((.....)))))((((((((((((((......))))))).)))))))..............((((((....))))))--.............---------- ( -31.30, z-score = -3.70, R) >droMoj3.scaffold_6540 25860372 99 + 34148556 UGAUGAUGAUGAUGGCCAUCAGCUGUCAACGGUUCUCACCCAAACCGUCUGACUGGGUAAAUAUUUACCAGCUGAGCAAUUGCAGC--AAGACGAAAAGAU---------- ..............((..((((((((((((((((........)))))).))))..(((((....)))))))))))((....)).))--.............---------- ( -26.30, z-score = -1.30, R) >consensus _UAAGGCGACGGCGACCAUCAGCUGUCAACAGCUUUGGCU______G_CUGUCUGCCUAAAUAUUUGCCAGCUGCGUAAUUGCAGCCGAAA_CUAAAGAGC_AAAA_____ ...((((......(((........)))...(((....)))..........))))................((((((....))))))......................... (-12.75 = -12.46 + -0.29)


| Location | 21,362,589 – 21,362,693 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 74.93 |
| Shannon entropy | 0.46407 |
| G+C content | 0.43658 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -14.58 |
| Energy contribution | -14.92 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
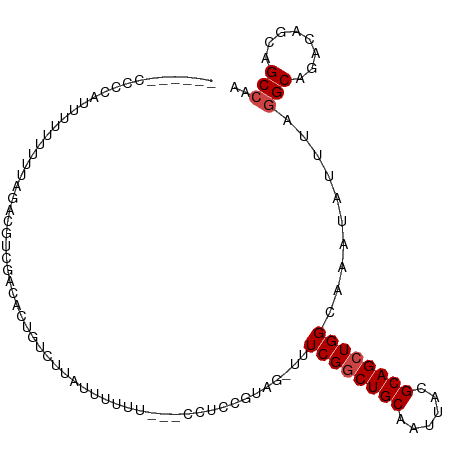
>dm3.chr3R 21362589 104 - 27905053 GCCUCUUUCUUUUUUUUUUUUUUUGUCGACUCUGUCUUAUUUUUU---GCUCCGUAG-UUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAGCAGCCAA ......................((((.(..(((((((.....(((---(((.((...-...))(((((......))))).)))))).....)))))))).)))).... ( -24.20, z-score = -1.64, R) >droSim1.chr3R 21190281 94 - 27517382 -----GCCUCAUUUUUUUU-----GUCGACACUGUCUUAUUUUUU---GCUCCGCAG-UUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAGCAGCCAA -----((((.......(((-----(((((.(((((..........---.....))))-).))((((((......)))))))))))).....))))............. ( -25.56, z-score = -1.72, R) >droSec1.super_13 149083 99 - 2104621 -----GCCUCAUUUUUUUUUUUUCGUCGACACUGUCUUAUUUUUU---GCUCCGUAG-UUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAGCAGCCAA -----..........................((((((.....(((---(((.((...-...))(((((......))))).)))))).....))))))........... ( -22.40, z-score = -1.10, R) >droYak2.chr3R 3713140 93 + 28832112 ------GCCGCUUUUUUUGUAGACACUG----U-CUUAUUUUUUU---CCACCGCAG-UUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAGCAGCCAA ------(((............((.((((----(-...........---.....))))-).))((((((......))))))))).........(((........))).. ( -24.29, z-score = -1.34, R) >droEre2.scaffold_4820 3788141 96 + 10470090 ------GCCUC-UUUUUUGUAGACACUAGCUCU-UUUUAUUUUUA---CCUCCCUAG-UUUCGCCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAGCAGCCAA ------(((..-.........((.(((((....-...........---.....))))-).))((.(((......))))).))).........(((........))).. ( -18.05, z-score = -0.11, R) >droAna3.scaffold_13340 3617823 88 - 23697760 --------------------ACCCUUCGACAGUGUCUCAUUUUUACUGCCUCUCUAGUUUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUACGCAGACAGCGGCAAA --------------------.......(((...)))..........((((......((((((((((((......)))))))).))))......((.....)))))).. ( -21.10, z-score = -1.02, R) >consensus ______CCCCAUUUUUUUUUAGACGUCGACACUGUCUUAUUUUUU___CCUCCGUAG_UUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAGCAGCCAA ............................................................((((((((......))))))))..........(((........))).. (-14.58 = -14.92 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:23 2011