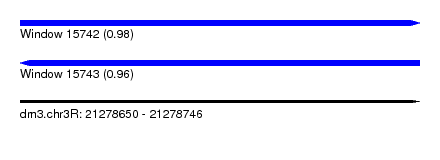
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,278,650 – 21,278,746 |
| Length | 96 |
| Max. P | 0.983892 |

| Location | 21,278,650 – 21,278,746 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 76.88 |
| Shannon entropy | 0.44506 |
| G+C content | 0.31948 |
| Mean single sequence MFE | -20.16 |
| Consensus MFE | -13.68 |
| Energy contribution | -14.19 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.983892 |
| Prediction | RNA |
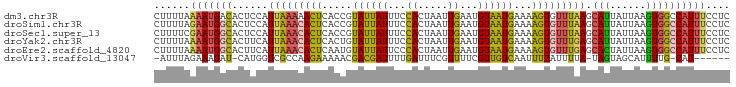
Download alignment: ClustalW | MAF
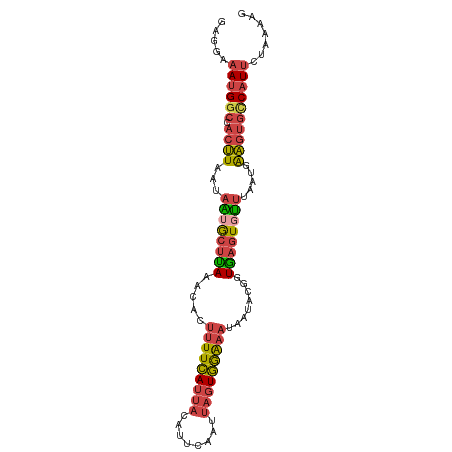
>dm3.chr3R 21278650 96 + 27905053 GAGGAAAUGGCCACUUAAUAAUGCUUAAACACUUUUCAUUACAUUCAAUUAGUGGAAAUAAUACGGUGAGUUUUUAAUGGAGUGUCAUUUUAAAAG ...((((((((.((((..(((.(((((.....(((((((((........)))))))))........)))))..)))...))))))))))))..... ( -18.62, z-score = -1.50, R) >droSim1.chr3R 21106711 96 + 27517382 GAGGAAAUGGCCACUUAAUAAUGCUUAAACACUUUUCAUUACAUUCAAUUAGUGGAAAUAAUACGGUGAGUGUUUAAUGGAGUGCCAUUCUAAAAG ..((((.((((.((((...((((((((.....(((((((((........)))))))))........)))))))).....))))))))))))..... ( -21.92, z-score = -2.09, R) >droSec1.super_13 64413 96 + 2104621 GAGGAAAUGGCCACUUAAUAAUGCUUAAACACUUUUCAUUACAUUCAAUUAGUGGAAAUAAUACGGUGAGUGUUUAAUGGAGUGCCAUUCGAAAAG ...(((.((((.((((...((((((((.....(((((((((........)))))))))........)))))))).....)))))))))))...... ( -21.02, z-score = -1.73, R) >droYak2.chr3R 3625706 96 - 28832112 GAGGAAAUGGCCACUUAAUAAUGCUCAAACACUUUUCAUUACAUUCAAUUAGUGGAAAUAAUACAGUGAGUGUUUAAUGAAGUGCCAUUUUAAAAG ...((((((((.((((...((((((((.....(((((((((........)))))))))........)))))))).....))))))))))))..... ( -25.32, z-score = -3.64, R) >droEre2.scaffold_4820 3703537 96 - 10470090 GAGGAAAUGGCCACUUAAUAGUGCUCAAACACUUUUCAUUACAUUCAAUUAGUGGGAAUAAUACAUUGAGUGUUUAAUGAAGUGCCAUUUUAAAAG ...((((((((.((((...((..(((((....((..(((((........)))))..)).......)))))..)).....))))))))))))..... ( -24.70, z-score = -3.08, R) >droVir3.scaffold_13047 2079286 86 + 19223366 ------AUG-CAAAAUGCUACUA-UAAAAUAAAAUUGACAACGAAAACGAAAUCAAAAUCGUCGUUUUUCUUGGCGACCAUG-AUCUUUCUAAAU- ------..(-(.....)).....-..........(((.(((.((((((((...........)))))))).))).))).....-............- ( -9.40, z-score = -0.17, R) >consensus GAGGAAAUGGCCACUUAAUAAUGCUUAAACACUUUUCAUUACAUUCAAUUAGUGGAAAUAAUACGGUGAGUGUUUAAUGAAGUGCCAUUCUAAAAG .....((((((.((((...((((((((.....(((((((((........)))))))))........)))))))).....))))))))))....... (-13.68 = -14.19 + 0.51)


| Location | 21,278,650 – 21,278,746 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Shannon entropy | 0.44506 |
| G+C content | 0.31948 |
| Mean single sequence MFE | -17.15 |
| Consensus MFE | -13.61 |
| Energy contribution | -12.78 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
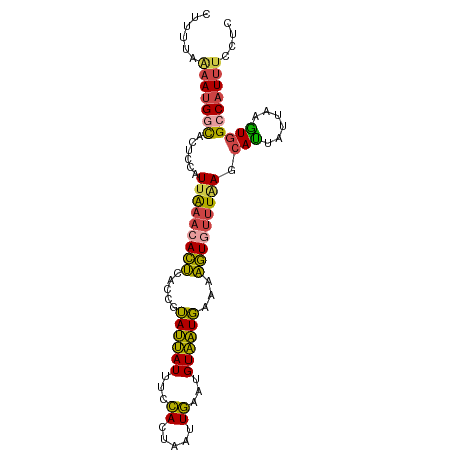
>dm3.chr3R 21278650 96 - 27905053 CUUUUAAAAUGACACUCCAUUAAAAACUCACCGUAUUAUUUCCACUAAUUGAAUGUAAUGAAAAGUGUUUAAGCAUUAUUAAGUGGCCAUUUCCUC ......(((((.((((.................(((((((((........))).))))))...(((((....)))))....))))..))))).... ( -9.60, z-score = 0.47, R) >droSim1.chr3R 21106711 96 - 27517382 CUUUUAGAAUGGCACUCCAUUAAACACUCACCGUAUUAUUUCCACUAAUUGAAUGUAAUGAAAAGUGUUUAAGCAUUAUUAAGUGGCCAUUUCCUC ......(((((((......(((((((((.....(((((((((........))).))))))...))))))))).((((....))))))))))).... ( -20.20, z-score = -2.58, R) >droSec1.super_13 64413 96 - 2104621 CUUUUCGAAUGGCACUCCAUUAAACACUCACCGUAUUAUUUCCACUAAUUGAAUGUAAUGAAAAGUGUUUAAGCAUUAUUAAGUGGCCAUUUCCUC ......(((((((......(((((((((.....(((((((((........))).))))))...))))))))).((((....))))))))))).... ( -20.20, z-score = -2.74, R) >droYak2.chr3R 3625706 96 + 28832112 CUUUUAAAAUGGCACUUCAUUAAACACUCACUGUAUUAUUUCCACUAAUUGAAUGUAAUGAAAAGUGUUUGAGCAUUAUUAAGUGGCCAUUUCCUC ......(((((((......(((((((((.....(((((((((........))).))))))...))))))))).((((....))))))))))).... ( -19.90, z-score = -2.18, R) >droEre2.scaffold_4820 3703537 96 + 10470090 CUUUUAAAAUGGCACUUCAUUAAACACUCAAUGUAUUAUUCCCACUAAUUGAAUGUAAUGAAAAGUGUUUGAGCACUAUUAAGUGGCCAUUUCCUC ......(((((((......(((((((((.....((((((...((.....))...))))))...))))))))).((((....))))))))))).... ( -21.60, z-score = -2.86, R) >droVir3.scaffold_13047 2079286 86 - 19223366 -AUUUAGAAAGAU-CAUGGUCGCCAAGAAAAACGACGAUUUUGAUUUCGUUUUCGUUGUCAAUUUUAUUUUA-UAGUAGCAUUUUG-CAU------ -...(((((.(((-....)))..........(((((((...((....))...)))))))...))))).....-.....((.....)-)..------ ( -11.40, z-score = 0.72, R) >consensus CUUUUAAAAUGGCACUCCAUUAAACACUCACCGUAUUAUUUCCACUAAUUGAAUGUAAUGAAAAGUGUUUAAGCAUUAUUAAGUGGCCAUUUCCUC ......(((((((......(((((((((.....((((((...((.....))...))))))...))))))))).(((......)))))))))).... (-13.61 = -12.78 + -0.83)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:15 2011